Exosomal and Non-Exosomal Transport of Extra-Cellular microRNAs in Follicular Fluid: Implications for Bovine Oocyte Developmental Competence
- PMID: 24223816
- PMCID: PMC3817212
- DOI: 10.1371/journal.pone.0078505
Exosomal and Non-Exosomal Transport of Extra-Cellular microRNAs in Follicular Fluid: Implications for Bovine Oocyte Developmental Competence
Abstract
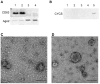
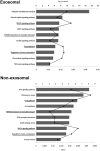
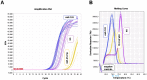
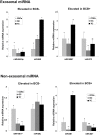
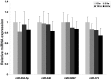
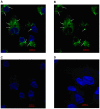
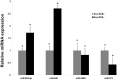
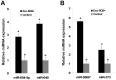
Cell-cell communication within the follicle involves many signaling molecules, and this process may be mediated by secretion and uptake of exosomes that contain several bioactive molecules including extra-cellular miRNAs. Follicular fluid and cells from individual follicles of cattle were grouped based on Brilliant Cresyl Blue (BCB) staining of the corresponding oocytes. Both Exoquick precipitation and differential ultracentrifugation were used to separate the exosome and non-exosomal fraction of follicular fluid. Following miRNA isolation from both fractions, the human miRCURY LNA™ Universal RT miRNA PCR array system was used to profile miRNA expression. This analysis found that miRNAs were present in both exosomal and non-exosomal fraction of bovine follicular fluid. We found 25 miRNAs differentially expressed (16 up and 9 down) in exosomes and 30 miRNAs differentially expressed (21 up and 9 down) in non-exosomal fraction of follicular fluid in comparison of BCB- versus BCB+ oocyte groups. Expression of selected miRNAs was detected in theca, granulosa and cumulus oocyte complex. To further explore the potential roles of these follicular fluid derived extra-cellular miRNAs, the potential target genes were predicted, and functional annotation and pathway analysis revealed most of these pathways are known regulators of follicular development and oocyte growth. In order to validate exosome mediated cell-cell communication within follicular microenvironment, we demonstrated uptake of exosomes and resulting increase of endogenous miRNA level and subsequent alteration of mRNA levels in follicular cells in vitro. This study demonstrates for the first time, the presence of exosome or non-exosome mediated transfer of miRNA in the bovine follicular fluid, and oocyte growth dependent variation in extra-cellular miRNA signatures in the follicular environment.
Conflict of interest statement
Figures
Similar articles
-
Prediction of major microRNAs in follicular fluid regulating porcine oocyte development.J Assist Reprod Genet. 2020 Oct;37(10):2569-2579. doi: 10.1007/s10815-020-01909-0. Epub 2020 Aug 11. J Assist Reprod Genet. 2020. PMID: 32780318 Free PMC article.
-
Regulation of ACVR1 and ID2 by cell-secreted exosomes during follicle maturation in the mare.Reprod Biol Endocrinol. 2014 May 26;12:44. doi: 10.1186/1477-7827-12-44. Reprod Biol Endocrinol. 2014. PMID: 24884710 Free PMC article.
-
The extent of the abundance of exosomal and non-exosomal extracellular miRNAs in the bovine follicular fluid.Reprod Domest Anim. 2022 Oct;57(10):1208-1217. doi: 10.1111/rda.14195. Epub 2022 Jul 8. Reprod Domest Anim. 2022. PMID: 35765751
-
Exosomal Communication Between Cumulus-Oocyte Complexes and Granulosa Cells: A New Molecular Axis for Oocyte Competence in Human-Assisted Reproduction.Int J Mol Sci. 2025 Jun 3;26(11):5363. doi: 10.3390/ijms26115363. Int J Mol Sci. 2025. PMID: 40508172 Free PMC article. Review.
-
The transcriptome of follicular cells: biological insights and clinical implications for the treatment of infertility.Hum Reprod Update. 2014 Jan-Feb;20(1):1-11. doi: 10.1093/humupd/dmt044. Epub 2013 Sep 29. Hum Reprod Update. 2014. PMID: 24082041 Free PMC article. Review.
Cited by
-
Extracellular vesicles affecting embryo development in vitro: a potential culture medium supplement.Front Pharmacol. 2024 Sep 18;15:1366992. doi: 10.3389/fphar.2024.1366992. eCollection 2024. Front Pharmacol. 2024. PMID: 39359245 Free PMC article. Review.
-
Seasonal influence on miRNA expression dynamics of extracellular vesicles in equine follicular fluid.J Anim Sci Biotechnol. 2024 Oct 9;15(1):137. doi: 10.1186/s40104-024-01097-2. J Anim Sci Biotechnol. 2024. PMID: 39380110 Free PMC article.
-
Diversity of Extracellular Vesicles in Human Follicular Fluid: Morphological Analysis and Quantification.Int J Mol Sci. 2022 Oct 2;23(19):11676. doi: 10.3390/ijms231911676. Int J Mol Sci. 2022. PMID: 36232981 Free PMC article.
-
Impact of NAD+ metabolism on ovarian aging.Immun Ageing. 2023 Dec 2;20(1):70. doi: 10.1186/s12979-023-00398-w. Immun Ageing. 2023. PMID: 38041117 Free PMC article. Review.
-
The role of extracellular vesicles in animal reproduction and diseases.J Anim Sci Biotechnol. 2022 Jun 10;13(1):62. doi: 10.1186/s40104-022-00715-1. J Anim Sci Biotechnol. 2022. PMID: 35681164 Free PMC article. Review.
References
-
- Eppig JJ (2001) Oocyte control of ovarian follicular development and function in mammals. Reproduction 122: 829–838. - PubMed
-
- Harwood BN, Cross SK, Radford EE, Haac BE, De Vries WN (2008) Members of the WNT signaling pathways are widely expressed in mouse ovaries, oocytes, and cleavage stage embryos. Dev Dyn 237: 1099–1111. - PubMed
-
- Su YQ, Wu X, O'Brien MJ, Pendola FL, Denegre JN, et al. (2004) Synergistic roles of BMP15 and GDF9 in the development and function of the oocyte-cumulus cell complex in mice: genetic evidence for an oocyte-granulosa cell regulatory loop. Dev Biol 276: 64–73. - PubMed
-
- Patsoula E, Loutradis D, Drakakis P, Kallianidis K, Bletsa R, et al. (2001) Expression of mRNA for the LH and FSH receptors in mouse oocytes and preimplantation embryos. Reproduction 121: 455–461. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

