SWEETLEAD: an in silico database of approved drugs, regulated chemicals, and herbal isolates for computer-aided drug discovery
- PMID: 24223973
- PMCID: PMC3815155
- DOI: 10.1371/journal.pone.0079568
SWEETLEAD: an in silico database of approved drugs, regulated chemicals, and herbal isolates for computer-aided drug discovery
Abstract
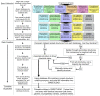
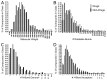
In the face of drastically rising drug discovery costs, strategies promising to reduce development timelines and expenditures are being pursued. Computer-aided virtual screening and repurposing approved drugs are two such strategies that have shown recent success. Herein, we report the creation of a highly-curated in silico database of chemical structures representing approved drugs, chemical isolates from traditional medicinal herbs, and regulated chemicals, termed the SWEETLEAD database. The motivation for SWEETLEAD stems from the observance of conflicting information in publicly available chemical databases and the lack of a highly curated database of chemical structures for the globally approved drugs. A consensus building scheme surveying information from several publicly accessible databases was employed to identify the correct structure for each chemical. Resulting structures are filtered for the active pharmaceutical ingredient, standardized, and differing formulations of the same drug were combined in the final database. The publically available release of SWEETLEAD (https://simtk.org/home/sweetlead) provides an important tool to enable the successful completion of computer-aided repurposing and drug discovery campaigns.
Conflict of interest statement
Figures


References
-
- Paul S, Mytelka D, Dunwiddie C, Persinger C, Munos B et al. (2010) How to improve R&D productivity: the pharmaceutical industry's grand challenge. Nat Rev Drug Discov: 12. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

