Bayesian phylogeography of Crimean-Congo hemorrhagic fever virus in Europe
- PMID: 24223988
- PMCID: PMC3817137
- DOI: 10.1371/journal.pone.0079663
Bayesian phylogeography of Crimean-Congo hemorrhagic fever virus in Europe
Abstract
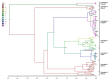
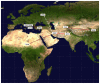
Crimean-Congo hemorrhagic fever (CCHF) is a zoonosis mainly transmitted by ticks that causes severe hemorrhagic fever and has a mortality rate of 5-60%. The first outbreak of CCHF occurred in the Crimean peninsula in 1944-45 and it has recently emerged in the Balkans and eastern Mediterranean. In order to reconstruct the origin and pathway of the worldwide dispersion of the virus at global and regional (eastern European) level, we investigated the phylogeography of the infection by analysing 121 publicly available CCHFV S gene sequences including two recently characterised Albanian isolates. The spatial and temporal phylogeny was reconstructed using a Bayesian Markov chain Monte Carlo approach, which estimated a mean evolutionary rate of 2.96 x 10(-4) (95%HPD=1.6 and 4.7 x 10(-4)) substitutions/site/year for the analysed fragment. All of the isolates segregated into seven highly significant clades that correspond to the known geographical clades: in particular the two new isolates from northern Albania clustered significantly within the Europe 1 clade. Our phylogeographical reconstruction suggests that the global CCHFV clades originated about one thousand years ago from a common ancestor probably located in Africa. The virus then spread to Asia in the XV century and entered Europe on at least two occasions: the first in the early 1800s, when a still circulating but less or non-pathogenic virus emerged in Greece and Turkey, and the second in the early 1900s, when a pathogenic CCHFV strain began to spread in eastern Europe. The most probable location for the origin of this European clade 1 was Russia, but Turkey played a central role in spreading the virus throughout Europe. Given the close proximity of the infected areas, our data suggest that the movement of wild and domestic ungulates from endemic areas was probably the main cause of the dissemination of the virus in eastern Europe.
Conflict of interest statement
Figures
References
-
- Schmaljohn CS, Hooper JW (2001) Bunyaviridae: the viruses and their replication. Fields virology.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

