Systems approaches evaluating the perturbation of xenobiotic metabolism in response to cigarette smoke exposure in nasal and bronchial tissues
- PMID: 24224167
- PMCID: PMC3808713
- DOI: 10.1155/2013/512086
Systems approaches evaluating the perturbation of xenobiotic metabolism in response to cigarette smoke exposure in nasal and bronchial tissues
Abstract
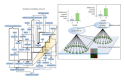
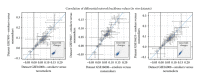
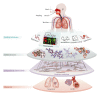
Capturing the effects of exposure in a specific target organ is a major challenge in risk assessment. Exposure to cigarette smoke (CS) implicates the field of tissue injury in the lung as well as nasal and airway epithelia. Xenobiotic metabolism in particular becomes an attractive tool for chemical risk assessment because of its responsiveness against toxic compounds, including those present in CS. This study describes an efficient integration from transcriptomic data to quantitative measures, which reflect the responses against xenobiotics that are captured in a biological network model. We show here that our novel systems approach can quantify the perturbation in the network model of xenobiotic metabolism. We further show that this approach efficiently compares the perturbation upon CS exposure in bronchial and nasal epithelial cells in vivo samples obtained from smokers. Our observation suggests the xenobiotic responses in the bronchial and nasal epithelial cells of smokers were similar to those observed in their respective organotypic models exposed to CS. Furthermore, the results suggest that nasal tissue is a reliable surrogate to measure xenobiotic responses in bronchial tissue.
Figures




References
-
- Sharma A, Saurabh K, Yadav S, Jain SK, Parmar D. Expression profiling of selected genes of toxication and detoxication pathways in peripheral blood lymphocytes as a biomarker for predicting toxicity of environmental chemicals. International Journal of Hygiene and Environmental Health. 2012 - PubMed
-
- Hecht SS. DNA adduct formation from tobacco-specific N-nitrosamines. Mutation Research. 1999;424(1-2):127–142. - PubMed
-
- Li D, Firozi PF, Wang L-E, et al. Sensitivity to DNA damage induced by benzo(a)pyrene diol epoxide and risk of lung cancer: a case-control analysis. Cancer Research. 2001;61(4):1445–1450. - PubMed
-
- Shimada T. Xenobiotic-metabolizing enzymes involved in activation and detoxification of carcinogenic polycyclic aromatic hydrocarbons. Drug Metabolism and Pharmacokinetics. 2006;21(4):257–276. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

