Elastic properties and secondary structure formation of single-stranded DNA at monovalent and divalent salt conditions
- PMID: 24225314
- PMCID: PMC3919573
- DOI: 10.1093/nar/gkt1089
Elastic properties and secondary structure formation of single-stranded DNA at monovalent and divalent salt conditions
Abstract
Single-stranded DNA (ssDNA) plays a major role in several biological processes. It is therefore of fundamental interest to understand how the elastic response and the formation of secondary structures are modulated by the interplay between base pairing and electrostatic interactions. Here we measure force-extension curves (FECs) of ssDNA molecules in optical tweezers set up over two orders of magnitude of monovalent and divalent salt conditions, and obtain its elastic parameters by fitting the FECs to semiflexible models of polymers. For both monovalent and divalent salts, we find that the electrostatic contribution to the persistence length is proportional to the Debye screening length, varying as the inverse of the square root of cation concentration. The intrinsic persistence length is equal to 0.7 nm for both types of salts, and the effectivity of divalent cations in screening electrostatic interactions appears to be 100-fold as compared with monovalent salt, in line with what has been recently reported for single-stranded RNA. Finally, we propose an analysis of the FECs using a model that accounts for the effective thickness of the filament at low salt condition and a simple phenomenological description that quantifies the formation of non-specific secondary structure at low forces.
Figures
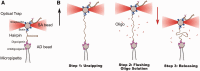
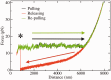
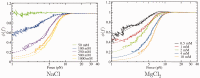
 1 pN) as a sudden jump-up in the force (highlighted with an asterisk). The re-pulling curve in the absence of blocking oligo is drawn in green/dark gray [black and green/dark gray curves nearly superimpose due to the quasi-reversibility of the unfolding-folding process, see [(25)].
1 pN) as a sudden jump-up in the force (highlighted with an asterisk). The re-pulling curve in the absence of blocking oligo is drawn in green/dark gray [black and green/dark gray curves nearly superimpose due to the quasi-reversibility of the unfolding-folding process, see [(25)].

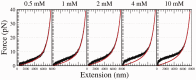
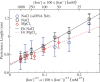
 =
= (0)+A*
(0)+A*  are:
are:  (0) = 0.68
(0) = 0.68  0.04 and A = 2.19
0.04 and A = 2.19  0.10 (
0.10 ( = 0.666) in the case of NaCl; and
= 0.666) in the case of NaCl; and  (0)=0.70
(0)=0.70  0.04 and A = 1.59
0.04 and A = 1.59  0.57 (
0.57 ( =0.701) in the case of MgCl2.
=0.701) in the case of MgCl2.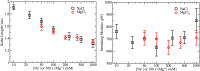

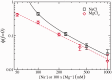
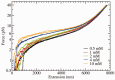
 at different salt concentrations. Logarithmic scale is used for both axes. Black squares stand for NaCl and red diamonds stand for MgCl2. Magnesium concentrations have been multiplied by a factor 100. Lines are fits to the power law dependence,
at different salt concentrations. Logarithmic scale is used for both axes. Black squares stand for NaCl and red diamonds stand for MgCl2. Magnesium concentrations have been multiplied by a factor 100. Lines are fits to the power law dependence,  (see text for details).
(see text for details).Similar articles
-
Stretching single-stranded DNA: interplay of electrostatic, base-pairing, and base-pair stacking interactions.Biophys J. 2001 Aug;81(2):1133-43. doi: 10.1016/S0006-3495(01)75770-0. Biophys J. 2001. PMID: 11463654 Free PMC article.
-
Sugar-Pucker Force-Induced Transition in Single-Stranded DNA.Int J Mol Sci. 2021 Apr 29;22(9):4745. doi: 10.3390/ijms22094745. Int J Mol Sci. 2021. PMID: 33947069 Free PMC article.
-
Sequence-dependent elasticity and electrostatics of single-stranded DNA: signatures of base-stacking.Biophys J. 2014 Feb 4;106(3):659-66. doi: 10.1016/j.bpj.2013.12.018. Biophys J. 2014. PMID: 24507606 Free PMC article.
-
DNA structure: cations in charge?Curr Opin Struct Biol. 1999 Jun;9(3):298-304. doi: 10.1016/S0959-440X(99)80040-2. Curr Opin Struct Biol. 1999. PMID: 10361089 Review.
-
Pyruvate kinase: activation by and catalytic role of the monovalent and divalent cations.Mol Cell Biochem. 1981 Mar 13;35(2):65-75. doi: 10.1007/BF02354821. Mol Cell Biochem. 1981. PMID: 7015112 Review.
Cited by
-
Temperature-dependent elastic properties of DNA.Biophys Rep (N Y). 2022 Aug 5;2(3):100067. doi: 10.1016/j.bpr.2022.100067. eCollection 2022 Sep 14. Biophys Rep (N Y). 2022. PMID: 36425333 Free PMC article.
-
Subangstrom single-molecule measurements of motor proteins using a nanopore.Nat Biotechnol. 2015 Oct;33(10):1073-5. doi: 10.1038/nbt.3357. Epub 2015 Sep 28. Nat Biotechnol. 2015. PMID: 26414351 Free PMC article.
-
Stacking correlation length in single-stranded DNA.Nucleic Acids Res. 2024 Nov 27;52(21):13243-13254. doi: 10.1093/nar/gkae934. Nucleic Acids Res. 2024. PMID: 39460618 Free PMC article.
-
Tension Gauge Tethers as Tension Threshold and Duration Sensors.ACS Sens. 2023 Feb 24;8(2):704-711. doi: 10.1021/acssensors.2c02218. Epub 2023 Feb 2. ACS Sens. 2023. PMID: 36731861 Free PMC article.
-
Templated folding of the RTX domain of the bacterial toxin adenylate cyclase revealed by single molecule force spectroscopy.Nat Commun. 2022 May 19;13(1):2784. doi: 10.1038/s41467-022-30448-8. Nat Commun. 2022. PMID: 35589788 Free PMC article.
References
-
- Saenger W. Principles of Nucleic Acid Structure. New York: Springer; 1984.
-
- Bloomfield VA, Crothers DM, Tinoco IJ. Nucleic Acids. Sausalito, CA, USA: University Science Books; 1999.
-
- Zhou H, Zhang Y. Pulling hairpinned polynucleotide chains: does base-pair stacking interaction matter? J. Chem. Phys. 2001;114:8694–8700.

