The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases
- PMID: 24225315
- PMCID: PMC3964957
- DOI: 10.1093/nar/gkt1103
The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases
Abstract
The MetaCyc database (MetaCyc.org) is a comprehensive and freely accessible database describing metabolic pathways and enzymes from all domains of life. MetaCyc pathways are experimentally determined, mostly small-molecule metabolic pathways and are curated from the primary scientific literature. MetaCyc contains >2100 pathways derived from >37,000 publications, and is the largest curated collection of metabolic pathways currently available. BioCyc (BioCyc.org) is a collection of >3000 organism-specific Pathway/Genome Databases (PGDBs), each containing the full genome and predicted metabolic network of one organism, including metabolites, enzymes, reactions, metabolic pathways, predicted operons, transport systems and pathway-hole fillers. Additions to BioCyc over the past 2 years include YeastCyc, a PGDB for Saccharomyces cerevisiae, and 891 new genomes from the Human Microbiome Project. The BioCyc Web site offers a variety of tools for querying and analysis of PGDBs, including Omics Viewers and tools for comparative analysis. New developments include atom mappings in reactions, a new representation of glycan degradation pathways, improved compound structure display, better coverage of enzyme kinetic data, enhancements of the Web Groups functionality, improvements to the Omics viewers, a new representation of the Enzyme Commission system and, for the desktop version of the software, the ability to save display states.
Figures
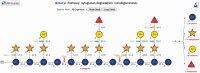


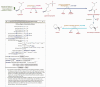
References
-
- Christie KR, Weng S, Balakrishnan R, Costanzo MC, Dolinski K, Dwight SS, Engel SR, Feierbach B, Fisk DG, Hirschman JE, et al. Saccharomyces genome database (SGD) provides tools to identify and analyze sequences from Saccharomyces cerevisiae and related sequences from other organisms. Nucleic Acids Res. 2004;32:D311–D314. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

