MicroRNAs transfer from human macrophages to hepato-carcinoma cells and inhibit proliferation
- PMID: 24227773
- PMCID: PMC3858238
- DOI: 10.4049/jimmunol.1301728
MicroRNAs transfer from human macrophages to hepato-carcinoma cells and inhibit proliferation
Abstract
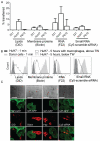
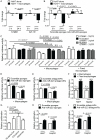
Recent research has indicated a new mode of intercellular communication facilitated by the movement of RNA between cells. There is evidence that RNA can transfer between cells in a multitude of ways, including in complex with proteins or lipids or in vesicles, including apoptotic bodies and exosomes. However, there remains little understanding of the function of nucleic acid transfer between human cells. In this article, we report that human macrophages transfer microRNAs (miRNAs) to hepato-carcinoma cells (HCCs) in a manner that required intercellular contact and involved gap junctions. Two specific miRNAs transferred efficiently between these cells--miR-142 and miR-223--and both were endogenously expressed in macrophages and not in HCCs. Transfer of these miRNAs influenced posttranscriptional regulation of proteins in HCCs, including decreased expression of reporter proteins and endogenously expressed stathmin-1 and insulin-like growth factor-1 receptor. Importantly, transfer of miRNAs from macrophages functionally inhibited proliferation of these cancerous cells. Thus, these data led us to propose that intercellular transfer of miRNA from immune cells could serve as a new defense against unwanted cell proliferation or tumor growth.
Figures



References
-
- Chitwood DH, Timmermans MCP. Small RNAs are on the move. Nature. 2010;467:415–419. - PubMed
-
- Zernecke A, Bidzhekov K, Noels H, Shagdarsuren E, Gan L, Denecke B, Hristov M, Köppel T, Jahantigh MN, Lutgens E, Wang S, Olson EN, Schober A, Weber C. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Science Signaling. 2009;2:ra81. - PubMed
-
- Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biology. 2007;9:654–659. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

