The Drosophila early ovarian transcriptome provides insight to the molecular causes of recombination rate variation across genomes
- PMID: 24228734
- PMCID: PMC3840681
- DOI: 10.1186/1471-2164-14-794
The Drosophila early ovarian transcriptome provides insight to the molecular causes of recombination rate variation across genomes
Abstract
Background: Evidence in yeast indicates that gene expression is correlated with recombination activity and double-strand break (DSB) formation in some hotspots. Studies of nucleosome occupancy in yeast and mice also suggest that open chromatin influences the formation of DSBs. In Drosophila melanogaster, high-resolution recombination maps show an excess of DSBs within annotated transcripts relative to intergenic sequences. The impact of active transcription on recombination landscapes, however, remains unexplored in a multicellular organism. We then investigated the transcription profile during early meiosis in D. melanogaster females to obtain a glimpse at the relevant transcriptional dynamics during DSB formation, and test the specific hypothesis that DSBs preferentially target transcriptionally active genomic regions.
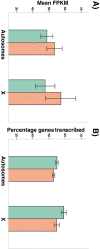
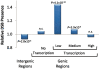
Results: Our study of transcript profiles of early- and late-meiosis using mRNA-seq revealed, 1) significant differences in gene expression, 2) new genes and exons, 3) parent-of-origin effects on transcription in early-meiosis stages, and 4) a nonrandom genomic distribution of transcribed genes. Importantly, genomic regions that are more actively transcribed during early meiosis show higher rates of recombination, and we ruled out DSB preference for genic regions that are not transcribed.
Conclusions: Our results provide evidence in a multicellular organism that transcription during the initial phases of meiosis increases the likelihood of DSB and give insight into the molecular determinants of recombination rate variation across the D. melanogaster genome. We propose that a model where variation in gene expression plays a role altering the recombination landscape across the genome could provide a molecular, heritable and plastic mechanism to observed patterns of recombination variation, from the high level of intra-specific variation to the known influence of environmental factors and stress conditions.
Figures

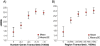
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

