ESET methylates UBF at K232/254 and regulates nucleolar heterochromatin plasticity and rDNA transcription
- PMID: 24234436
- PMCID: PMC3919562
- DOI: 10.1093/nar/gkt1041
ESET methylates UBF at K232/254 and regulates nucleolar heterochromatin plasticity and rDNA transcription
Abstract
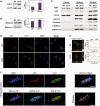
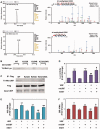
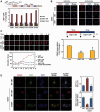
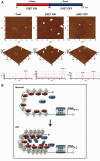
The remodeling of chromatin in the nucleolus is important for the control of ribosomal DNA (rDNA) transcription and ribosome biogenesis. Herein, we found that upstream binding factor (UBF) interacts with ESET, a histone H3K9 methyltransferase and is trimethylated at Lys (K) 232/254 by ESET. UBF trimethylation leads to nucleolar chromatin condensation and decreased rDNA transcriptional activity. UBF mutations at K232/254A and K232/254R restored rDNA transcriptional activity in response to ESET. Both ESET-ΔSET mutant and knockdown of ESET by short hairpin RNA reduced trimethylation of UBF and resulted in the restoration of rDNA transcription. Atomic force microscopy confirmed that UBF trimethylated by ESET modulates the plasticity of nucleolar chromatin. We further demonstrated that UBF trimethylation at K232/254 by ESET deregulates rDNA transcription in a cell model of Huntington's disease. Together, our findings show that a novel epigenetic modification of UBF is linked to impaired rDNA transcription and nucleolar chromatin remodeling, which may play key roles in the pathogenesis of neurodegeneration.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

