Functional microRNAs and target sites are created by lineage-specific transposition
- PMID: 24234653
- PMCID: PMC3943519
- DOI: 10.1093/hmg/ddt569
Functional microRNAs and target sites are created by lineage-specific transposition
Abstract
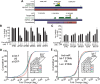
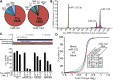
Transposable elements (TEs) account for nearly one-half of the sequence content in the human genome, and de novo germline transposition into regulatory or coding sequences of protein-coding genes can cause heritable disorders. TEs are prevalent in and around protein-coding genes, providing an opportunity to impart regulation. Computational studies reveal that microRNA (miRNA) genes and miRNA target sites reside within TE sequences, but there is little experimental evidence supporting a role for TEs in the birth of miRNAs, or as platform for gene regulation by miRNAs. In this work, we validate miRNAs and target sites derived from TE families prevalent in the human genome, including the ancient long interspersed nuclear element 2 (LINE2/L2), mammalian-wide interspersed repeat (MIR) retrotransposons and the primate-specific Alu family. We show that genes with 3' untranslated region (3' UTR) MIR elements are enriched for let-7 targets and that these sites are conserved and responsive to let-7 expression. We also demonstrate that 3' UTR-embedded Alus are a source of miR-24 and miR-122 target sites and that a subset of active genomic Alus provide for de novo target site creation. Finally, we report that although the creation of miRNA genes by Alu elements is relatively uncommon relative to their overall genomic abundance, Alu-derived miR-1285-1 is efficiently processed from its genomic locus and regulates genes with target sites contained within homologous elements. Taken together, our data provide additional evidence for TEs as a source for miRNAs and miRNA target sites, with instances of conservation through the course of mammalian evolution.
Figures



References
-
- Xing J., Witherspoon D.J., Ray D.A., Batzer M.A., Jorde L.B. Mobile DNA elements in primate and human evolution. Am J Phys Anthropol. 2007 Suppl 45, 2–19. - PubMed
-
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

