Automethylation activities within the mixed lineage leukemia-1 (MLL1) core complex reveal evidence supporting a "two-active site" model for multiple histone H3 lysine 4 methylation
- PMID: 24235145
- PMCID: PMC3887211
- DOI: 10.1074/jbc.M113.501064
Automethylation activities within the mixed lineage leukemia-1 (MLL1) core complex reveal evidence supporting a "two-active site" model for multiple histone H3 lysine 4 methylation
Abstract
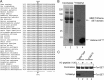
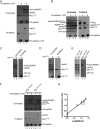
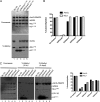
The mixed lineage leukemia-1 (MLL1) core complex predominantly catalyzes mono- and dimethylation of histone H3 at lysine 4 (H3K4) and is frequently altered in aggressive acute leukemias. The molecular mechanisms that account for conversion of mono- to dimethyl H3K4 (H3K4me1,2) are not well understood. In this investigation, we report that the suppressor of variegation, enhancer of zeste, trithorax (SET) domains from human MLL1 and Drosophila Trithorax undergo robust intramolecular automethylation reactions at an evolutionarily conserved cysteine residue in the active site, which is inhibited by unmodified histone H3. The location of the automethylation in the SET-I subdomain indicates that the MLL1 SET domain possesses significantly more conformational plasticity in solution than suggested by its crystal structure. We also report that MLL1 methylates Ash2L in the absence of histone H3, but only when assembled within a complex with WDR5 and RbBP5, suggesting a restraint for the architectural arrangement of subunits within the complex. Using MLL1 and Ash2L automethylation reactions as probes for histone binding, we observed that both automethylation reactions are significantly inhibited by stoichiometric amounts of unmethylated histone H3, but not by histones previously mono-, di-, or trimethylated at H3K4. These results suggest that the H3K4me1 intermediate does not significantly bind to the MLL1 SET domain during the dimethylation reaction. Consistent with this hypothesis, we demonstrate that the MLL1 core complex assembled with a catalytically inactive SET domain variant preferentially catalyzes H3K4 dimethylation using the H3K4me1 substrate. Taken together, these results are consistent with a "two-active site" model for multiple H3K4 methylation by the MLL1 core complex.
Keywords: Automethylation; Chromatin Histone Modification; Enzyme Mechanisms; Epigenetics; Histone Methylation; Leukemia; Self-methylation.
Figures





Similar articles
-
On the mechanism of multiple lysine methylation by the human mixed lineage leukemia protein-1 (MLL1) core complex.J Biol Chem. 2009 Sep 4;284(36):24242-56. doi: 10.1074/jbc.M109.014498. Epub 2009 Jun 25. J Biol Chem. 2009. PMID: 19556245 Free PMC article.
-
An Ash2L/RbBP5 heterodimer stimulates the MLL1 methyltransferase activity through coordinated substrate interactions with the MLL1 SET domain.PLoS One. 2010 Nov 23;5(11):e14102. doi: 10.1371/journal.pone.0014102. PLoS One. 2010. PMID: 21124902 Free PMC article.
-
Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.J Biol Chem. 2012 Aug 10;287(33):27275-89. doi: 10.1074/jbc.M112.364125. Epub 2012 Jun 3. J Biol Chem. 2012. PMID: 22665483 Free PMC article.
-
The Development of Inhibitors Targeting the Mixed Lineage Leukemia 1 (MLL1)-WD Repeat Domain 5 Protein (WDR5) Protein- Protein Interaction.Curr Med Chem. 2020;27(33):5530-5542. doi: 10.2174/0929867326666190528080514. Curr Med Chem. 2020. PMID: 31132972 Review.
-
Mixed lineage leukemia: a structure-function perspective of the MLL1 protein.FEBS J. 2010 Apr;277(8):1832-42. doi: 10.1111/j.1742-4658.2010.07609.x. Epub 2010 Mar 4. FEBS J. 2010. PMID: 20236310 Free PMC article. Review.
Cited by
-
Biochemical reconstitution and phylogenetic comparison of human SET1 family core complexes involved in histone methylation.J Biol Chem. 2015 Mar 6;290(10):6361-75. doi: 10.1074/jbc.M114.627646. Epub 2015 Jan 5. J Biol Chem. 2015. PMID: 25561738 Free PMC article.
-
Hierarchical assembly of the MLL1 core complex regulates H3K4 methylation and is dependent on temperature and component concentration.J Biol Chem. 2023 Feb;299(2):102874. doi: 10.1016/j.jbc.2023.102874. Epub 2023 Jan 6. J Biol Chem. 2023. PMID: 36623730 Free PMC article.
-
Characterization of SETD3 methyltransferase-mediated protein methionine methylation.J Biol Chem. 2020 Aug 7;295(32):10901-10910. doi: 10.1074/jbc.RA120.014072. Epub 2020 Jun 5. J Biol Chem. 2020. PMID: 32503840 Free PMC article.
-
The histone methyltransferase activity of MLL1 is dispensable for hematopoiesis and leukemogenesis.Cell Rep. 2014 May 22;7(4):1239-47. doi: 10.1016/j.celrep.2014.04.015. Epub 2014 May 9. Cell Rep. 2014. PMID: 24813891 Free PMC article.
-
Investigation of the methylation of Numb by the SET8 protein lysine methyltransferase.Sci Rep. 2015 Sep 22;5:13813. doi: 10.1038/srep13813. Sci Rep. 2015. PMID: 26391684 Free PMC article.
References
-
- Djabali M., Selleri L., Parry P., Bower M., Young B. D., Evans G. A. (1992) A trithorax-like gene is interrupted by chromosome 11q23 translocations in acute leukaemias. Nat. Genet. 2, 113–118 - PubMed
-
- Gu Y., Nakamura T., Alder H., Prasad R., Canaani O., Cimino G., Croce C. M., Canaani E. (1992) The t(4;11) chromosome translocation of human acute leukemias fuses the ALL-1 gene, related to Drosophila trithorax, to the AF-4 gene. Cell 71, 701–708 - PubMed
-
- Tkachuk D. C., Kohler S., Cleary M. L. (1992) Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell 71, 691–700 - PubMed
-
- Ziemin-van der Poel S., McCabe N. R., Gill H. J., Espinosa R., 3rd, Patel Y., Harden A., Rubinelli P., Smith S. D., LeBeau M. M., Rowley J. D., et al. (1991) Identification of a gene, MLL, that spans the breakpoint in 11q23 translocations associated with human leukemias. Proc. Natl. Acad. Sci. U.S.A. 88, 10735–10739 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

