Pediatric fecal microbiota harbor diverse and novel antibiotic resistance genes
- PMID: 24236055
- PMCID: PMC3827270
- DOI: 10.1371/journal.pone.0078822
Pediatric fecal microbiota harbor diverse and novel antibiotic resistance genes
Abstract
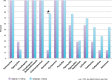
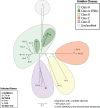
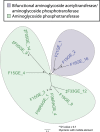
Emerging antibiotic resistance threatens human health. Gut microbes are an epidemiologically important reservoir of resistance genes (resistome), yet prior studies indicate that the true diversity of gut-associated resistomes has been underestimated. To deeply characterize the pediatric gut-associated resistome, we created metagenomic recombinant libraries in an Escherichia coli host using fecal DNA from 22 healthy infants and children (most without recent antibiotic exposure), and performed functional selections for resistance to 18 antibiotics from eight drug classes. Resistance-conferring DNA fragments were sequenced (Illumina HiSeq 2000), and reads assembled and annotated with the PARFuMS computational pipeline. Resistance to 14 of the 18 antibiotics was found in stools of infants and children. Recovered genes included chloramphenicol acetyltransferases, drug-resistant dihydrofolate reductases, rRNA methyltransferases, transcriptional regulators, multidrug efflux pumps, and every major class of beta-lactamase, aminoglycoside-modifying enzyme, and tetracycline resistance protein. Many resistance-conferring sequences were mobilizable; some had low identity to any known organism, emphasizing cryptic organisms as potentially important resistance reservoirs. We functionally confirmed three novel resistance genes, including a 16S rRNA methylase conferring aminoglycoside resistance, and two tetracycline-resistance proteins nearly identical to a bifidobacterial MFS transporter (B. longum s. longum JDM301). We provide the first report to our knowledge of resistance to folate-synthesis inhibitors conferred by a predicted Nudix hydrolase (part of the folate synthesis pathway). This functional metagenomic survey of gut-associated resistomes, the largest of its kind to date, demonstrates that fecal resistomes of healthy children are far more diverse than previously suspected, that clinically relevant resistance genes are present even without recent selective antibiotic pressure in the human host, and that cryptic gut microbes are an important resistance reservoir. The observed transferability of gut-associated resistance genes to a gram-negative (E. coli) host also suggests that the potential for gut-associated resistomes to threaten human health by mediating antibiotic resistance in pathogens warrants further investigation.
Conflict of interest statement
Figures
References
-
- Alekshun MN, Levy SB (2007) Molecular mechanisms of antibacterial multidrug resistance. Cell 128: 1037–1050. - PubMed
-
- Robicsek A, Jacoby GA, Hooper DC (2006) The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect Dis 6: 629–640. - PubMed
-
- Salyers AA, Gupta A, Wang Y (2004) Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol 12: 412–416. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

