Molecular cloning and characterization of porcine Na⁺/K⁺-ATPase isoforms α1, α2, α3 and the ATP1A3 promoter
- PMID: 24236096
- PMCID: PMC3827302
- DOI: 10.1371/journal.pone.0079127
Molecular cloning and characterization of porcine Na⁺/K⁺-ATPase isoforms α1, α2, α3 and the ATP1A3 promoter
Abstract
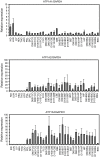
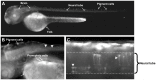
Na⁺/K⁺-ATPase maintains electrochemical gradients of Na⁺ and K⁺ essential for a variety of cellular functions including neuronal activity. The α-subunit of the Na⁺/K⁺-ATPase exists in four different isoforms (α1-α4) encoded by different genes. With a view to future use of pig as an animal model in studies of human diseases caused by Na⁺/K⁺-ATPase mutations, we have determined the porcine coding sequences of the α1-α3 genes, ATP1A1, ATP1A2, and ATP1A3, their chromosomal localization, and expression patterns. Our ATP1A1 sequence accords with the sequences from several species at five positions where the amino acid residue of the previously published porcine ATP1A1 sequence differs. These corrections include replacement of glutamine 841 with arginine. Analysis of the functional consequences of substitution of the arginine revealed its importance for Na⁺ binding, which can be explained by interaction of the arginine with the C-terminus, stabilizing one of the Na⁺ sites. Quantitative real-time PCR expression analyses of porcine ATP1A1, ATP1A2, and ATP1A3 mRNA showed that all three transcripts are expressed in the embryonic brain as early as 60 days of gestation. Expression of α3 is confined to neuronal tissue. Generally, the expression patterns of ATP1A1, ATP1A2, and ATP1A3 transcripts were found similar to their human counterparts, except for lack of α3 expression in porcine heart. These expression patterns were confirmed at the protein level. We also report the sequence of the porcine ATP1A3 promoter, which was found to be closely homologous to its human counterpart. The function and specificity of the porcine ATP1A3 promoter was analyzed in transgenic zebrafish, demonstrating that it is active and drives expression in embryonic brain and spinal cord. The results of the present study provide a sound basis for employing the ATP1A3 promoter in attempts to generate transgenic porcine models of neurological diseases caused by ATP1A3 mutations.
Conflict of interest statement
Figures








References
-
- Skou JC (1957) The influence of some cations on an adenosine triphosphatase from peripheral nerves. Biochim Biophys Acta 23: 394–401. - PubMed
-
- Post RL, Hegyvary C, Kume S (1972) Activation by adenosine triphosphate in the phosphorylation kinetics of sodium and potassium ion transport adenosine triphosphatase. J Biol Chem 247: 6530–6540. - PubMed
-
- Morth JP, Pedersen BP, Toustrup-Jensen MS, Sorensen TL, Petersen J, et al. (2007) Crystal structure of the sodium-potassium pump. Nature 450: 1043–1049. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

