Genetic variation of human papillomavirus type 16 in individual clinical specimens revealed by deep sequencing
- PMID: 24236186
- PMCID: PMC3827439
- DOI: 10.1371/journal.pone.0080583
Genetic variation of human papillomavirus type 16 in individual clinical specimens revealed by deep sequencing
Erratum in
- PLoS One. 2014;9(1). doi:10.1371/annotation/524fa93c-e77c-4c3d-bf6e-763a5b057409
Abstract
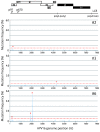
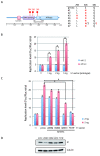
Viral genetic diversity within infected cells or tissues, called viral quasispecies, has been mostly studied for RNA viruses, but has also been described among DNA viruses, including human papillomavirus type 16 (HPV16) present in cervical precancerous lesions. However, the extent of HPV genetic variation in cervical specimens, and its involvement in HPV-induced carcinogenesis, remains unclear. Here, we employ deep sequencing to comprehensively analyze genetic variation in the HPV16 genome isolated from individual clinical specimens. Through overlapping full-circle PCR, approximately 8-kb DNA fragments covering the whole HPV16 genome were amplified from HPV16-positive cervical exfoliated cells collected from patients with either low-grade squamous intraepithelial lesion (LSIL) or invasive cervical cancer (ICC). Deep sequencing of the amplified HPV16 DNA enabled de novo assembly of the full-length HPV16 genome sequence for each of 7 specimens (5 LSIL and 2 ICC samples). Subsequent alignment of read sequences to the assembled HPV16 sequence revealed that 2 LSILs and 1 ICC contained nucleotide variations within E6, E1 and the non-coding region between E5 and L2 with mutation frequencies of 0.60% to 5.42%. In transient replication assays, a novel E1 mutant found in ICC, E1 Q381E, showed reduced ability to support HPV16 origin-dependent replication. In addition, partially deleted E2 genes were detected in 1 LSIL sample in a mixed state with the intact E2 gene. Thus, the methods used in this study provide a fundamental framework for investigating the influence of HPV somatic genetic variation on cervical carcinogenesis.
Conflict of interest statement
Figures




Similar articles
-
Within-Host Variations of Human Papillomavirus Reveal APOBEC Signature Mutagenesis in the Viral Genome.J Virol. 2018 May 29;92(12):e00017-18. doi: 10.1128/JVI.00017-18. Print 2018 Jun 15. J Virol. 2018. PMID: 29593040 Free PMC article.
-
Association of human papillomavirus type 16 long control region mutation and cervical cancer.Virol J. 2013 Jan 23;10:30. doi: 10.1186/1743-422X-10-30. Virol J. 2013. PMID: 23343096 Free PMC article.
-
HPV prevalence, E6 sequence variation and physical state of HPV16 isolates from patients with cervical cancer in Sichuan, China.Gynecol Oncol. 2007 Jan;104(1):77-85. doi: 10.1016/j.ygyno.2006.07.016. Epub 2006 Sep 12. Gynecol Oncol. 2007. PMID: 16970982
-
Genetic variations of human papillomavirus type 16: implications for cervical carcinogenesis.Jpn J Infect Dis. 2015;68(3):169-75. doi: 10.7883/yoken.JJID.2014.584. Epub 2015 Mar 13. Jpn J Infect Dis. 2015. PMID: 25766614 Review.
-
Genetic variability of the HPV16 early genes and LCR. Present and future perspectives.Expert Rev Mol Med. 2021 Dec 1;23:e19. doi: 10.1017/erm.2021.18. Expert Rev Mol Med. 2021. PMID: 34847982 Review.
Cited by
-
Whole Genome Sequencing of A(H3N2) Influenza Viruses Reveals Variants Associated with Severity during the 2016⁻2017 Season.Viruses. 2019 Jan 28;11(2):108. doi: 10.3390/v11020108. Viruses. 2019. PMID: 30695992 Free PMC article.
-
Whole-genome analysis of human papillomavirus genotypes 52 and 58 isolated from Japanese women with cervical intraepithelial neoplasia and invasive cervical cancer.Infect Agent Cancer. 2017 Aug 4;12:44. doi: 10.1186/s13027-017-0155-4. eCollection 2017. Infect Agent Cancer. 2017. PMID: 28785305 Free PMC article.
-
Ancient Evolution and Dispersion of Human Papillomavirus 58 Variants.J Virol. 2017 Oct 13;91(21):e01285-17. doi: 10.1128/JVI.01285-17. Print 2017 Nov 1. J Virol. 2017. PMID: 28794033 Free PMC article.
-
Characterization of Intra-Type Variants of Oncogenic Human Papillomaviruses by Next-Generation Deep Sequencing of the E6/E7 Region.Viruses. 2016 Mar 14;8(3):79. doi: 10.3390/v8030079. Viruses. 2016. PMID: 26985902 Free PMC article.
-
TaME-seq: An efficient sequencing approach for characterisation of HPV genomic variability and chromosomal integration.Sci Rep. 2019 Jan 24;9(1):524. doi: 10.1038/s41598-018-36669-6. Sci Rep. 2019. PMID: 30679491 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

