Prediction and characterization of small non-coding RNAs related to secondary metabolites in Saccharopolyspora erythraea
- PMID: 24236194
- PMCID: PMC3827479
- DOI: 10.1371/journal.pone.0080676
Prediction and characterization of small non-coding RNAs related to secondary metabolites in Saccharopolyspora erythraea
Abstract
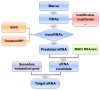
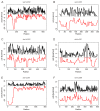
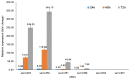
Saccharopolyspora erythraea produces a large number of secondary metabolites with biological activities, including erythromycin. Elucidation of the mechanisms through which the production of these secondary metabolites is regulated may help to identify new strategies for improved biosynthesis of erythromycin. In this paper, we describe the systematic prediction and analysis of small non-coding RNAs (sRNAs) in S. erythraea, with the aim to elucidate sRNA-mediated regulation of secondary metabolite biosynthesis. In silico and deep-sequencing technologies were applied to predict sRNAs in S. erythraea. Six hundred and forty-seven potential sRNA loci were identified, of which 382 cis-encoded antisense RNA are complementary to protein-coding regions and 265 predicted transcripts are located in intergenic regions. Six candidate sRNAs (sernc292, sernc293, sernc350, sernc351, sernc361, and sernc389) belong to four gene clusters (tpc3, pke, pks6, and nrps5) that are involved in secondary metabolite biosynthesis. Deep-sequencing data showed that the expression of all sRNAs in the strain HL3168 E3 (E3) was higher than that in NRRL23338 (M), except for sernc292 and sernc361 expression. The relative expression of six sRNAs in strain M and E3 were validated by qRT-PCR at three different time points (24, 48, and 72 h). The results showed that, at each time point, the transcription levels of sernc293, sernc350, sernc351, and sernc389 were higher in E3 than in M, with the largest difference observed at 72 h, whereas no signals for sernc292 and sernc361 were detected. sernc293, sernc350, sernc351, and sernc389 probably regulate iron transport, terpene metabolism, geosmin synthesis, and polyketide biosynthesis, respectively. The major significance of this study is the successful prediction and identification of sRNAs in genomic regions close to the secondary metabolism-related genes in S. erythraea. A better understanding of the sRNA-target interaction would help to elucidate the complete range of functions of sRNAs in S. erythraea, including sRNA-mediated regulation of erythromycin biosynthesis.
Conflict of interest statement
Figures

Similar articles
-
Systems perspectives on erythromycin biosynthesis by comparative genomic and transcriptomic analyses of S. erythraea E3 and NRRL23338 strains.BMC Genomics. 2013 Jul 31;14:523. doi: 10.1186/1471-2164-14-523. BMC Genomics. 2013. PMID: 23902230 Free PMC article.
-
SACE_5599, a putative regulatory protein, is involved in morphological differentiation and erythromycin production in Saccharopolyspora erythraea.Microb Cell Fact. 2013 Dec 17;12:126. doi: 10.1186/1475-2859-12-126. Microb Cell Fact. 2013. PMID: 24341557 Free PMC article.
-
DasR is a pleiotropic regulator required for antibiotic production, pigment biosynthesis, and morphological development in Saccharopolyspora erythraea.Appl Microbiol Biotechnol. 2015 Dec;99(23):10215-24. doi: 10.1007/s00253-015-6892-7. Epub 2015 Aug 14. Appl Microbiol Biotechnol. 2015. PMID: 26272095
-
Small RNA-mediated regulation in bacteria: A growing palette of diverse mechanisms.Gene. 2018 May 20;656:60-72. doi: 10.1016/j.gene.2018.02.068. Epub 2018 Mar 1. Gene. 2018. PMID: 29501814 Review.
-
[Biogenesis and regulation of biosynthesis of erythromycins in Saccharopolyspora erythraea: a review].Prikl Biokhim Mikrobiol. 2004 Nov-Dec;40(6):613-24. Prikl Biokhim Mikrobiol. 2004. PMID: 15609849 Review. Russian.
Cited by
-
Synthetic biology approaches to actinomycete strain improvement.FEMS Microbiol Lett. 2021 Jun 11;368(10):fnab060. doi: 10.1093/femsle/fnab060. FEMS Microbiol Lett. 2021. PMID: 34057181 Free PMC article. Review.
-
Identification of Small RNAs in Streptomyces clavuligerus Using High-Resolution Transcriptomics and Expression Profiling During Clavulanic Acid Production.Int J Mol Sci. 2024 Dec 16;25(24):13472. doi: 10.3390/ijms252413472. Int J Mol Sci. 2024. PMID: 39769236 Free PMC article.
-
Omics for Bioprospecting and Drug Discovery from Bacteria and Microalgae.Antibiotics (Basel). 2020 May 4;9(5):229. doi: 10.3390/antibiotics9050229. Antibiotics (Basel). 2020. PMID: 32375367 Free PMC article. Review.
-
Coupled Transcriptomics for Differential Expression Analysis and Determination of Transcription Start Sites: Design and Bioinformatics.Methods Mol Biol. 2021;2296:263-278. doi: 10.1007/978-1-0716-1358-0_16. Methods Mol Biol. 2021. PMID: 33977454
References
-
- LABEDA DP (1987) Transfer of the Type Strain of Streptomyces erythraeus (Waksman 1923) Waksman and Henrici 1948 to the Genus Saccharopolyspora Lacey and Goodfellow 1975 as Saccharopolyspora erythraea sp. nov., and Designation of a Neotype Strain for Streptomyces erythraeus. Int J Syst Bacteriol 37: 19-22. doi:10.1099/00207713-37-1-19. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

