Processing of damaged DNA ends for double-strand break repair in mammalian cells
- PMID: 24236237
- PMCID: PMC3825254
- DOI: 10.5402/2012/345805
Processing of damaged DNA ends for double-strand break repair in mammalian cells
Abstract
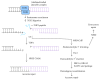
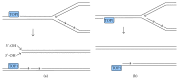
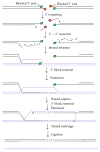
Most DNA double-strand breaks (DSBs)formed in a natural environment have chemical modifications at or near the ends that preclude direct religation and require removal or other processing so that rejoining can proceed. Free radical-mediated DSBs typically bear unligatable 3'-phosphate or 3'-phosphoglycolate termini and often have oxidized bases and/or abasic sites near the break. Topoisomerase-mediated DSBs are blocked by covalently bound peptide fragments of the topoisomerase. Enzymes capable of resolving damaged ends include polynucleotide kinase/phosphatase, which restores missing 5'-phosphates and removes 3'-phosphates; tyrosyl-DNA phosphodiesterases I and II (TDP1 and TDP2), which remove peptide fragments of topoisomerases I and II, respectively, and the Artemis and Metnase endonucleases, which can trim damaged overhangs of diverse structure. TDP1 as well as APE1 can remove 3'-phosphoglycolates and other 3' blocks, while CtIP appears to provide an alternative pathway for topoisomerase II fragment removal. Ku, a core DSB joining protein, can cleave abasic sites near DNA ends. The downstream processes of patching and ligation are tolerant of residual damage, and can sometimes proceed without complete damage removal. Despite these redundant pathways for resolution, damaged ends appear to be a significant barrier to rejoining, and their resolution may be a rate-limiting step in repair of some DSBs..
Figures




References
-
- Dedon P. C., Goldberg I. H. Free-radical mechanisms involved in the formation of sequence-dependent bistranded DNA lesions by the antitumor antibiotics bleomycin, neocarzinostatin, and calicheamicin. Chemical Research in Toxicology. 1992;5(3):311–332. - PubMed
-
- Iliakis G. E., Pantelias G. E., Okayasu R., Blakely W. F. Induction by H2O2 of DNA and interphase chromosome damage in plateau- phase Chinese hamster ovary cells. Radiation Research. 1992;131(2):192–203. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
