Crystal structure of Vδ1 T cell receptor in complex with CD1d-sulfatide shows MHC-like recognition of a self-lipid by human γδ T cells
- PMID: 24239091
- PMCID: PMC3875342
- DOI: 10.1016/j.immuni.2013.11.001
Crystal structure of Vδ1 T cell receptor in complex with CD1d-sulfatide shows MHC-like recognition of a self-lipid by human γδ T cells
Abstract
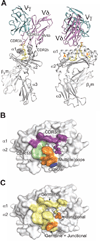
The nature of the antigens recognized by γδ T cells and their potential recognition of major histocompatibility complex (MHC)-like molecules has remained unclear. Members of the CD1 family of lipid-presenting molecules are suggested ligands for Vδ1 TCR-expressing γδ T cells, the major γδ lymphocyte population in epithelial tissues. We crystallized a Vδ1 TCR in complex with CD1d and the self-lipid sulfatide, revealing the unusual recognition of CD1d by germline Vδ1 residues spanning all complementarity-determining region (CDR) loops, as well as sulfatide recognition separately encoded by nongermline CDR3δ residues. Binding and functional analysis showed that CD1d presenting self-lipids, including sulfatide, was widely recognized by gut Vδ1+ γδ T cells. These findings provide structural demonstration of MHC-like recognition of a self-lipid by γδ T cells and reveal the prevalence of lipid recognition by innate-like T cell populations.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures





Comment in
-
A long-playing CD about the γδ TCR repertoire.Immunity. 2013 Dec 12;39(6):994-6. doi: 10.1016/j.immuni.2013.11.016. Immunity. 2013. PMID: 24332025
References
-
- Adams EJ, Chien YH, Garcia KC. Structure of a gammadelta T cell receptor in complex with the nonclassical MHC T22. Science. 2005;308:227–231. - PubMed
-
- Bauer S, Groh V, Wu J, Steinle A, Phillips JH, Lanier LL, Spies T. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science. 1999;285:727–729. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

