A comparative study of Pointed and Yan expression reveals new complexity to the transcriptional networks downstream of receptor tyrosine kinase signaling
- PMID: 24240101
- PMCID: PMC3947276
- DOI: 10.1016/j.ydbio.2013.11.002
A comparative study of Pointed and Yan expression reveals new complexity to the transcriptional networks downstream of receptor tyrosine kinase signaling
Abstract
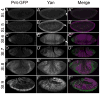
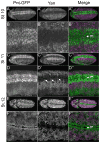
The biochemical regulatory network downstream of receptor tyrosine kinase (RTK) signaling is controlled by two opposing ETS family members: the transcriptional activator Pointed (Pnt) and the transcriptional repressor Yan. A bistable switch model has been invoked to explain how pathway activation can drive differentiation by shifting the system from a high-Yan/low-Pnt activity state to a low-Yan/high-Pnt activity state. Although the model explains yan and pnt loss-of-function phenotypes in several different cell types, how Yan and Pointed protein expression dynamics contribute to these and other developmental transitions remains poorly understood. Toward this goal we have used a functional GFP-tagged Pnt transgene (Pnt-GFP) to perform a comparative study of Yan and Pnt protein expression throughout Drosophila development. Consistent with the prevailing model of the Pnt-Yan network, we found numerous instances where Pnt-GFP and Yan adopt a mutually exclusive pattern of expression. However we also observed many examples of co-expression. While some co-expression occurred in cells where RTK signaling is presumed low, other co-expression occurred in cells with high RTK signaling. The instances of co-expressed Yan and Pnt-GFP in tissues with high RTK signaling cannot be explained by the current model, and thus they provide important contexts for future investigation of how context-specific differences in RTK signaling, network topology, or responsiveness to other signaling inputs, affect the transcriptional response.
Keywords: Drosophila; EGFR signaling; ETS transcription factor; Embryo; Gene regulation; Imaginal discs.
© 2013 Published by Elsevier Inc.
Figures









References
-
- Affolter M, Itoh N, Shilo B, Thiery J, Werb Z. Tube or Not Tube : Remodeling Epithelial Tissues by Branching Morphogenesis Branching morphogenesis involves the restructuring. Development. 2003;4:11–18. - PubMed
-
- Amin Ghabrial SL, Metzstein Mark M, Krasnow MA. BRANCHING MORPHOGENESIS OF THE DROSOPHILA TRACHEAL SYSTEM 2003 - PubMed
-
- Blair SS. Wing vein patterning in Drosophila and the analysis of intercellular signaling. Annual review of cell and developmental biology. 2007;23:293–319. - PubMed
-
- Boisclair Lachance JF, Fregoso Lomas M, Eleiche A, Bouchard Kerr P, Nilson LA. Graded Egfr activity patterns the Drosophila eggshell independently of autocrine feedback. Development Cambridge England. 2009;136:2893–2902. - PubMed
-
- Brunner D, Dücker K, Oellers N, Hafen E, Scholz H, Klämbt C. The ETS domain protein pointed-P2 is a target of MAP kinase in the sevenless signal transduction pathway. Nature. 1994;370:386–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

