Transcriptome sequencing during mouse brain development identifies long non-coding RNAs functionally involved in neurogenic commitment
- PMID: 24240175
- PMCID: PMC3981144
- DOI: 10.1038/emboj.2013.245
Transcriptome sequencing during mouse brain development identifies long non-coding RNAs functionally involved in neurogenic commitment
Abstract
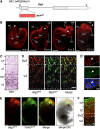
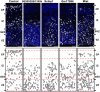
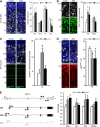
Transcriptome analysis of somatic stem cells and their progeny is fundamental to identify new factors controlling proliferation versus differentiation during tissue formation. Here, we generated a combinatorial, fluorescent reporter mouse line to isolate proliferating neural stem cells, differentiating progenitors and newborn neurons that coexist as intermingled cell populations during brain development. Transcriptome sequencing revealed numerous novel long non-coding (lnc)RNAs and uncharacterized protein-coding transcripts identifying the signature of neurogenic commitment. Importantly, most lncRNAs overlapped neurogenic genes and shared with them a nearly identical expression pattern suggesting that lncRNAs control corticogenesis by tuning the expression of nearby cell fate determinants. We assessed the power of our approach by manipulating lncRNAs and protein-coding transcripts with no function in corticogenesis reported to date. This led to several evident phenotypes in neurogenic commitment and neuronal survival, indicating that our study provides a remarkably high number of uncharacterized transcripts with hitherto unsuspected roles in brain development. Finally, we focussed on one lncRNA, Miat, whose manipulation was found to trigger pleiotropic effects on brain development and aberrant splicing of Wnt7b. Hence, our study suggests that lncRNA-mediated alternative splicing of cell fate determinants controls stem-cell commitment during neurogenesis.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

