Deorphanizing the human transmembrane genome: A landscape of uncharacterized membrane proteins
- PMID: 24241348
- PMCID: PMC3880479
- DOI: 10.1038/aps.2013.142
Deorphanizing the human transmembrane genome: A landscape of uncharacterized membrane proteins
Abstract
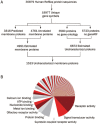
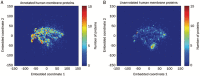
The sequencing of the human genome has fueled the last decade of work to functionally characterize genome content. An important subset of genes encodes membrane proteins, which are the targets of many drugs. They reside in lipid bilayers, restricting their endogenous activity to a relatively specialized biochemical environment. Without a reference phenotype, the application of systematic screens to profile candidate membrane proteins is not immediately possible. Bioinformatics has begun to show its effectiveness in focusing the functional characterization of orphan proteins of a particular functional class, such as channels or receptors. Here we discuss integration of experimental and bioinformatics approaches for characterizing the orphan membrane proteome. By analyzing the human genome, a landscape reference for the human transmembrane genome is provided.
Figures



References
-
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
-
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science. 2001;291:1304–51. - PubMed
-
- Fagerberg L, Jonasson K, von Heijne G, Uhlen M, Berglund L. Prediction of the human membrane proteome. Proteomics. 2010;10:1141–9. - PubMed
-
- Bakheet TM, Doig AJ. Properties and identification of human protein drug targets. Bioinformatics. 2009;25:451–7. - PubMed
-
- Yildirim MA, Goh KI, Cusick ME, Barabasi AL, Vidal M. Drug-target network. Nat Biotechnol. 2007;25:1119–26. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

