Strain-specific proteogenomics accelerates the discovery of natural products via their biosynthetic pathways
- PMID: 24242000
- PMCID: PMC3946956
- DOI: 10.1007/s10295-013-1373-4
Strain-specific proteogenomics accelerates the discovery of natural products via their biosynthetic pathways
Abstract
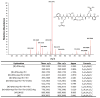
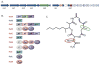
The use of proteomics for direct detection of expressed pathways producing natural products has yielded many new compounds, even when used in a screening mode without a bacterial genome sequence available. Here we quantify the advantages of having draft DNA-sequence available for strain-specific proteomics using the latest in ultrahigh-resolution mass spectrometry for both proteins and the small molecules they generate. Using the draft sequence of Streptomyces lilacinus NRRL B-1968, we show a >tenfold increase in the number of peptide identifications vs. using publicly available databases. Detected in this strain were six expressed gene clusters with varying homology to those known. To date, we have identified three of these clusters as encoding for the production of griseobactin (known), rakicidin D (an orphan NRPS/PKS hybrid cluster), and a putative thr and DHB-containing siderophore produced by a new non-ribosomal peptide sythetase gene cluster. The remaining three clusters show lower homology to those known, and likely encode enzymes for production of novel compounds. Using an interpreted strain-specific DNA sequence enables deep proteomics for the detection of multiple pathways and their encoded natural products in a single cultured bacterium.
Figures

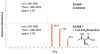

References
-
- Antibase: the natural compound identifier. Wiley VCH; 2011.
-
- Challis GL. Genome mining for novel natural product discovery. J Med Chem. 2008;51 (9):2618–2628. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

