Mg(2+)-induced conformational changes in the btuB riboswitch from E. coli
- PMID: 24243114
- PMCID: PMC3866643
- DOI: 10.1261/rna.039909.113
Mg(2+)-induced conformational changes in the btuB riboswitch from E. coli
Abstract
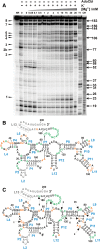
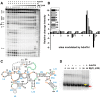
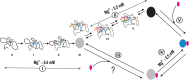
Mg(2+) has been shown to modulate the function of riboswitches by facilitating the ligand-riboswitch interactions. The btuB riboswitch from Escherichia coli undergoes a conformational change upon binding to its ligand, coenzyme B12 (adenosyl-cobalamine, AdoCbl), and down-regulates the expression of the B12 transporter protein BtuB in order to control the cellular levels of AdoCbl. Here, we discuss the structural folding attained by the btuB riboswitch from E. coli in response to Mg(2+) and how it affects the ligand binding competent conformation of the RNA. The btuB riboswitch notably adopts different conformational states depending upon the concentration of Mg(2+). With the help of in-line probing, we show the existence of at least two specific conformations, one being achieved in the complete absence of Mg(2+) (or low Mg(2+) concentration) and the other appearing above ∼0.5 mM Mg(2+). Distinct regions of the riboswitch exhibit different dissociation constants toward Mg(2+), indicating a stepwise folding of the btuB RNA. Increasing the Mg(2+) concentration drives the transition from one conformation toward the other. The conformational state existing above 0.5 mM Mg(2+) defines the binding competent conformation of the btuB riboswitch which can productively interact with the ligand, coenzyme B12, and switch the RNA conformation. Moreover, raising the Mg(2+) concentration enhances the ratio of switched RNA in the presence of AdoCbl. The lack of a AdoCbl-induced conformational switch experienced by the btuB riboswitch in the absence of Mg(2+) indicates a crucial role played by Mg(2+) for defining an active conformation of the riboswitch.
Keywords: Mg2+; RNA; coenzyme B12; folding; riboswitch.
Figures



Similar articles
-
A kissing loop is important for btuB riboswitch ligand sensing and regulatory control.J Biol Chem. 2015 Oct 30;290(44):26739-51. doi: 10.1074/jbc.M115.684134. Epub 2015 Sep 14. J Biol Chem. 2015. PMID: 26370077 Free PMC article.
-
The AdoCbl-riboswitch interaction investigated by in-line probing and surface plasmon resonance spectroscopy (SPR).Methods Enzymol. 2014;549:467-88. doi: 10.1016/B978-0-12-801122-5.00020-9. Methods Enzymol. 2014. PMID: 25432761
-
Corrin Ring Modifications Reveal the Chemical and Spatial Requirements for the B12-btuB Riboswitch Interaction.Chemistry. 2024 Sep 2;30(49):e202401800. doi: 10.1002/chem.202401800. Epub 2024 Aug 13. Chemistry. 2024. PMID: 38922714
-
A switch in time: detailing the life of a riboswitch.Biochim Biophys Acta. 2009 Sep-Oct;1789(9-10):584-91. doi: 10.1016/j.bbagrm.2009.06.004. Epub 2009 Jul 9. Biochim Biophys Acta. 2009. PMID: 19595806 Free PMC article. Review.
-
Alternative RNA Conformations: Companion or Combatant.Genes (Basel). 2022 Oct 23;13(11):1930. doi: 10.3390/genes13111930. Genes (Basel). 2022. PMID: 36360167 Free PMC article. Review.
Cited by
-
Context-dependence of T-loop Mediated Long-range RNA Tertiary Interactions.J Mol Biol. 2023 May 15;435(10):168070. doi: 10.1016/j.jmb.2023.168070. Epub 2023 Mar 31. J Mol Biol. 2023. PMID: 37003469 Free PMC article.
-
Tetracycline determines the conformation of its aptamer at physiological magnesium concentrations.Biophys J. 2014 Dec 16;107(12):2962-2971. doi: 10.1016/j.bpj.2014.11.001. Biophys J. 2014. PMID: 25517161 Free PMC article.
-
Structural and dynamic mechanisms for coupled folding and tRNA recognition of a translational T-box riboswitch.Nat Commun. 2023 Nov 15;14(1):7394. doi: 10.1038/s41467-023-43232-z. Nat Commun. 2023. PMID: 37968328 Free PMC article.
-
Tb3+-Cleavage Assays Reveal Specific Mg2+ Binding Sites Necessary to Pre-fold the btuB Riboswitch for AdoCbl Binding.Front Chem. 2017 Mar 21;5:10. doi: 10.3389/fchem.2017.00010. eCollection 2017. Front Chem. 2017. PMID: 28377919 Free PMC article.
-
Folding behavior of a T-shaped, ribosome-binding translation enhancer implicated in a wide-spread conformational switch.Elife. 2017 Feb 13;6:e22883. doi: 10.7554/eLife.22883. Elife. 2017. PMID: 28186489 Free PMC article.
References
-
- Bastet L, Dube A, Masse E, Lafontaine DA 2011. New insights into riboswitch regulation mechanisms. Mol Microbiol 80: 1148–1154 - PubMed
-
- Batey RT, Gilbert SD, Montange RK 2004. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature 432: 411–415 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
