Modeling integrated cellular machinery using hybrid Petri-Boolean networks
- PMID: 24244124
- PMCID: PMC3820535
- DOI: 10.1371/journal.pcbi.1003306
Modeling integrated cellular machinery using hybrid Petri-Boolean networks
Abstract
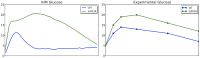
The behavior and phenotypic changes of cells are governed by a cellular circuitry that represents a set of biochemical reactions. Based on biological functions, this circuitry is divided into three types of networks, each encoding for a major biological process: signal transduction, transcription regulation, and metabolism. This division has generally enabled taming computational complexity dealing with the entire system, allowed for using modeling techniques that are specific to each of the components, and achieved separation of the different time scales at which reactions in each of the three networks occur. Nonetheless, with this division comes loss of information and power needed to elucidate certain cellular phenomena. Within the cell, these three types of networks work in tandem, and each produces signals and/or substances that are used by the others to process information and operate normally. Therefore, computational techniques for modeling integrated cellular machinery are needed. In this work, we propose an integrated hybrid model (IHM) that combines Petri nets and Boolean networks to model integrated cellular networks. Coupled with a stochastic simulation mechanism, the model simulates the dynamics of the integrated network, and can be perturbed to generate testable hypotheses. Our model is qualitative and is mostly built upon knowledge from the literature and requires fine-tuning of very few parameters. We validated our model on two systems: the transcriptional regulation of glucose metabolism in human cells, and cellular osmoregulation in S. cerevisiae. The model produced results that are in very good agreement with experimental data, and produces valid hypotheses. The abstract nature of our model and the ease of its construction makes it a very good candidate for modeling integrated networks from qualitative data. The results it produces can guide the practitioner to zoom into components and interconnections and investigate them using such more detailed mathematical models.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
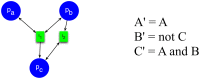
 ,
,  and
and  , where
, where  is self-regulatory (activating),
is self-regulatory (activating),  inhibits
inhibits  , and both
, and both  and
and  activate
activate  in a cooperative manner. (Left) A Petri net representation, with three places corresponding to the molecular species, and two transitions corresponding to the reactions. A read arc (line with arrows on both ends) connecting place
in a cooperative manner. (Left) A Petri net representation, with three places corresponding to the molecular species, and two transitions corresponding to the reactions. A read arc (line with arrows on both ends) connecting place  to transition
to transition  means that when transition
means that when transition  fires, the number of tokens in place
fires, the number of tokens in place  does not change. Notice that the inhibition of
does not change. Notice that the inhibition of  is represented by transition
is represented by transition  which consumes tokens from
which consumes tokens from  . (Right) A Boolean network representation, with three Boolean variables corresponding to the molecular species. The primed version of a variable indicated the next-state of that variable. In other words, these Boolean formulas can be interpreted a
. (Right) A Boolean network representation, with three Boolean variables corresponding to the molecular species. The primed version of a variable indicated the next-state of that variable. In other words, these Boolean formulas can be interpreted a  ,
,  , and
, and  .
.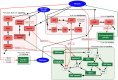
 , is set as follows: all Petri net places have 0 tokens except ADP (10 tokens) and Glucose (20 tokens); all Boolean network elements are set to 0, except HNF3beta and HNF1beta, which are set to 1. The ‘
, is set as follows: all Petri net places have 0 tokens except ADP (10 tokens) and Glucose (20 tokens); all Boolean network elements are set to 0, except HNF3beta and HNF1beta, which are set to 1. The ‘ ’ connections into Boolean variables correspond to the negation functions. For the Petri net component, the ‘
’ connections into Boolean variables correspond to the negation functions. For the Petri net component, the ‘ ’ connection from transition
’ connection from transition  to place
to place  is a schematic representation of inhibition, which is implemented using the standard Petri net definition as
is a schematic representation of inhibition, which is implemented using the standard Petri net definition as  being an input place to transition
being an input place to transition  . Transitions without inputs or outputs represent sources and sinks, respectively.
. Transitions without inputs or outputs represent sources and sinks, respectively.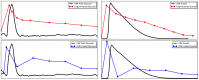
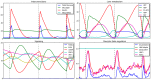


 , is set as follows: all Petri net places have 0 tokens except ADP, which has 10 tokens; all Boolean network elements are set to 0. See caption of Figure 2 for more details about the representation.
, is set as follows: all Petri net places have 0 tokens except ADP, which has 10 tokens; all Boolean network elements are set to 0. See caption of Figure 2 for more details about the representation.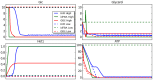

References
-
- Desvergne B, Michalik L, Wahli W (2006) Transcriptional regulation of metabolism. Physical Reviews 86: 465–514. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

