Removal of misincorporated ribonucleotides from prokaryotic genomes: an unexpected role for nucleotide excision repair
- PMID: 24244177
- PMCID: PMC3820734
- DOI: 10.1371/journal.pgen.1003878
Removal of misincorporated ribonucleotides from prokaryotic genomes: an unexpected role for nucleotide excision repair
Abstract
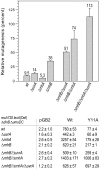
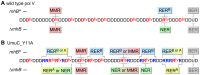
Stringent steric exclusion mechanisms limit the misincorporation of ribonucleotides by high-fidelity DNA polymerases into genomic DNA. In contrast, low-fidelity Escherichia coli DNA polymerase V (pol V) has relatively poor sugar discrimination and frequently misincorporates ribonucleotides. Substitution of a steric gate tyrosine residue with alanine (umuC_Y11A) reduces sugar selectivity further and allows pol V to readily misincorporate ribonucleotides as easily as deoxynucleotides, whilst leaving its poor base-substitution fidelity essentially unchanged. However, the mutability of cells expressing the steric gate pol V mutant is very low due to efficient repair mechanisms that are triggered by the misincorporated rNMPs. Comparison of the mutation frequency between strains expressing wild-type and mutant pol V therefore allows us to identify pathways specifically directed at ribonucleotide excision repair (RER). We previously demonstrated that rNMPs incorporated by umuC_Y11A are efficiently removed from DNA in a repair pathway initiated by RNase HII. Using the same approach, we show here that mismatch repair and base excision repair play minimal back-up roles in RER in vivo. In contrast, in the absence of functional RNase HII, umuC_Y11A-dependent mutagenesis increases significantly in ΔuvrA, uvrB5 and ΔuvrC strains, suggesting that rNMPs misincorporated into DNA are actively repaired by nucleotide excision repair (NER) in vivo. Participation of NER in RER was confirmed by reconstituting ribonucleotide-dependent NER in vitro. We show that UvrABC nuclease-catalyzed incisions are readily made on DNA templates containing one, two, or five rNMPs and that the reactions are stimulated by the presence of mispaired bases. Similar to NER of DNA lesions, excision of rNMPs proceeds through dual incisions made at the 8(th) phosphodiester bond 5' and 4(th)-5(th) phosphodiester bonds 3' of the ribonucleotide. Ribonucleotides misinserted into DNA can therefore be added to the broad list of helix-distorting modifications that are substrates for NER.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Lindahl T (1993) Instability and decay of the primary structure of DNA. Nature 362: 709–715. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

