Activin signaling targeted by insulin/dFOXO regulates aging and muscle proteostasis in Drosophila
- PMID: 24244197
- PMCID: PMC3820802
- DOI: 10.1371/journal.pgen.1003941
Activin signaling targeted by insulin/dFOXO regulates aging and muscle proteostasis in Drosophila
Abstract
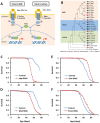
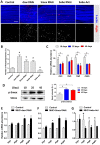
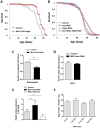
Reduced insulin/IGF signaling increases lifespan in many animals. To understand how insulin/IGF mediates lifespan in Drosophila, we performed chromatin immunoprecipitation-sequencing analysis with the insulin/IGF regulated transcription factor dFOXO in long-lived insulin/IGF signaling genotypes. Dawdle, an Activin ligand, is bound and repressed by dFOXO when reduced insulin/IGF extends lifespan. Reduced Activin signaling improves performance and protein homeostasis in muscles of aged flies. Activin signaling through the Smad binding element inhibits the transcription of Autophagy-specific gene 8a (Atg8a) within muscle, a factor controlling the rate of autophagy. Expression of Atg8a within muscle is sufficient to increase lifespan. These data reveal how insulin signaling can regulate aging through control of Activin signaling that in turn controls autophagy, representing a potentially conserved molecular basis for longevity assurance. While reduced Activin within muscle autonomously retards functional aging of this tissue, these effects in muscle also reduce secretion of insulin-like peptides at a distance from the brain. Reduced insulin secretion from the brain may subsequently reinforce longevity assurance through decreased systemic insulin/IGF signaling.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Tatar M, Bartke A, Antebi A (2003) The endocrine regulation of aging by insulin-like signals. Science 299: 1346–1351. - PubMed
-
- Kenyon C (2010) The genetics of ageing. Nature 464: 504–512. - PubMed
-
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R (1993) A C. elegans mutant that lives twice as long as wild type. Nature 366: 461–464. - PubMed
-
- Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G (1997) daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans . Science 277: 942–946. - PubMed
-
- Clancy DJ, Gems D, Harshman LG, Oldham S, Stocker H, et al. (2001) Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science 292: 104–106. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

