Transcriptional activity, chromosomal distribution and expression effects of transposable elements in Coffea genomes
- PMID: 24244387
- PMCID: PMC3823963
- DOI: 10.1371/journal.pone.0078931
Transcriptional activity, chromosomal distribution and expression effects of transposable elements in Coffea genomes
Abstract
Plant genomes are massively invaded by transposable elements (TEs), many of which are located near host genes and can thus impact gene expression. In flowering plants, TE expression can be activated (de-repressed) under certain stressful conditions, both biotic and abiotic, as well as by genome stress caused by hybridization. In this study, we examined the effects of these stress agents on TE expression in two diploid species of coffee, Coffea canephora and C. eugenioides, and their allotetraploid hybrid C. arabica. We also explored the relationship of TE repression mechanisms to host gene regulation via the effects of exonized TE sequences. Similar to what has been seen for other plants, overall TE expression levels are low in Coffea plant cultivars, consistent with the existence of effective TE repression mechanisms. TE expression patterns are highly dynamic across the species and conditions assayed here are unrelated to their classification at the level of TE class or family. In contrast to previous results, cell culture conditions per se do not lead to the de-repression of TE expression in C. arabica. Results obtained here indicate that differing plant drought stress levels relate strongly to TE repression mechanisms. TEs tend to be expressed at significantly higher levels in non-irrigated samples for the drought tolerant cultivars but in drought sensitive cultivars the opposite pattern was shown with irrigated samples showing significantly higher TE expression. Thus, TE genome repression mechanisms may be finely tuned to the ideal growth and/or regulatory conditions of the specific plant cultivars in which they are active. Analysis of TE expression levels in cell culture conditions underscored the importance of nonsense-mediated mRNA decay (NMD) pathways in the repression of Coffea TEs. These same NMD mechanisms can also regulate plant host gene expression via the repression of genes that bear exonized TE sequences.
Conflict of interest statement
Figures
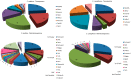

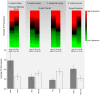


References
-
- Jordan IK, Rogozin IB, Glazko GV, Koonin EV (2003) Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet 19: 68–72. - PubMed
-
- Thornburg BG, Gotea V, Makalowski W (2006) Transposable elements as a significant source of transcription regulating signals. Gene 365: 104–110. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

