Comparative transcriptome analysis of salivary glands of two populations of rice brown planthopper, Nilaparvata lugens, that differ in virulence
- PMID: 24244529
- PMCID: PMC3828371
- DOI: 10.1371/journal.pone.0079612
Comparative transcriptome analysis of salivary glands of two populations of rice brown planthopper, Nilaparvata lugens, that differ in virulence
Abstract
Background: The brown planthopper (BPH), Nilaparvata lugens (Stål), a destructive rice pest in Asia, can quickly overcome rice resistance by evolving new virulent populations. Herbivore saliva plays an important role in plant-herbivore interactions, including in plant defense and herbivore virulence. However, thus far little is known about BPH saliva at the molecular level, especially its role in virulence and BPH-rice interaction.
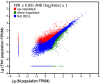
Methodology/principal findings: Using cDNA amplification in combination with Illumina short-read sequencing technology, we sequenced the salivary-gland transcriptomes of two BPH populations with different virulence; the populations were derived from rice variety TN1 (TN1 population) and Mudgo (M population). In total, 37,666 and 38,451 unigenes were generated from the salivary glands of these populations, respectively. When combined, a total of 43,312 unigenes were obtained, about 18 times more than the number of expressed sequence tags previously identified from these glands. Gene ontology annotations and KEGG orthology classifications indicated that genes related to metabolism, binding and transport were significantly active in the salivary glands. A total of 352 genes were predicted to encode secretory proteins, and some might play important roles in BPH feeding and BPH-rice interactions. Comparative analysis of the transcriptomes of the two populations revealed that the genes related to 'metabolism,' 'digestion and absorption,' and 'salivary secretion' might be associated with virulence. Moreover, 67 genes encoding putative secreted proteins were differentially expressed between the two populations, suggesting these genes may contribute to the change in virulence.
Conclusions/significance: This study was the first to compare the salivary-gland transcriptomes of two BPH populations having different virulence traits and to find genes that may be related to this difference. Our data provide a rich molecular resource for future functional studies on salivary glands and will be useful for elucidating the molecular mechanisms underlying BPH feeding and virulence differences.
Conflict of interest statement
Figures





References
-
- Miles PW (1999) Aphid saliva. Biol Rev 74: 41–85.
-
- DE Vos M, Jander G (2009) Myzus persicae (green peach aphid) salivary components induce defence responses in Arabidopsis thaliana . Plant Cell Environ 32: 1548–1560. - PubMed
-
- Hogenhout SA, Bos JIB (2011) Effector proteins that modulate plant-insect interactions. Curr Opin Plant Biol 14: 422–428. - PubMed
-
- Nicholson SJ, Hartson SD, Puterka GJ (2012) Proteomic analysis of secreted saliva from Russian Wheat Aphid (Diuraphis noxia Kurd.) biotypes that differ in virulence to wheat. J Proteomics 75: 2252–2268. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

