Identification of significant features in DNA microarray data
- PMID: 24244802
- PMCID: PMC3826574
- DOI: 10.1002/wics.1260
Identification of significant features in DNA microarray data
Abstract
DNA microarrays are a relatively new technology that can simultaneously measure the expression level of thousands of genes. They have become an important tool for a wide variety of biological experiments. One of the most common goals of DNA microarray experiments is to identify genes associated with biological processes of interest. Conventional statistical tests often produce poor results when applied to microarray data owing to small sample sizes, noisy data, and correlation among the expression levels of the genes. Thus, novel statistical methods are needed to identify significant genes in DNA microarray experiments. This article discusses the challenges inherent in DNA microarray analysis and describes a series of statistical techniques that can be used to overcome these challenges. The problem of multiple hypothesis testing and its relation to microarray studies are also considered, along with several possible solutions.
Keywords: feature selection; genetics; microarray; multiple testing.
Figures


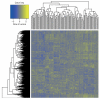
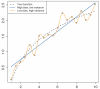
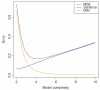
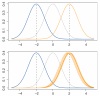
References
-
- International Human Genome Sequencing Consortium Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. - PubMed
-
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
-
- Dudoit S, Yang Y, Callow MJ, Speed TP. Statistical methods for identifying differentially expressed genes in replicated cDNA microarray experiments. Stat Sin. 2002;12:111–139.
-
- Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, Mittmann M, Wang C, Kobayashi M, Horton H, et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996;14:1675–1680. - PubMed
-
- DeRisi J, Penland L, Brown PO, Bittner ML, Meltzer PS, Ray M, Chen Y, Su YA, Trent JM. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat Genet. 1996;14:457–460. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
