Design and analysis of a tunable synchronized oscillator
- PMID: 24245660
- PMCID: PMC4177200
- DOI: 10.1186/1754-1611-7-26
Design and analysis of a tunable synchronized oscillator
Abstract
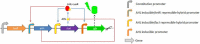
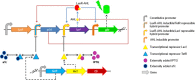
Background: The use of in silico simulations as a basis for designing artificial biological systems (and experiments to characterize them) is one of the tangible differences between Synthetic Biology and "classical" Genetic Engineering. To this end, synthetic biologists have adopted approaches originating from the traditionally non-biological fields of Nonlinear Dynamics and Systems & Control Theory. However, due to the complex molecular interactions affecting the emergent properties of biological systems, mechanistic descriptions of even the simplest genetic circuits (transcriptional feedback oscillators, bi-stable switches) produced by these methods tend to be either oversimplified, or numerically intractable. More comprehensive and realistic models can be approximated by constructing "toy" genetic circuits that provide the experimenter with some degree of control over the transcriptional dynamics, and allow for experimental set-ups that generate reliable data reflecting the intracellular biochemical state in real time. To this end, we designed two genetic circuits (basic and tunable) capable of exhibiting synchronized oscillatory green fluorescent protein (GFP) expression in small populations of Escherichia coli cells. The functionality of the basic circuit was verified microscopically. High-level visualizations of computational simulations were analyzed to determine whether the reliability and utility of a synchronized transcriptional oscillator could be enhanced by the introduction of chemically inducible repressors.
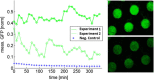
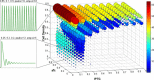
Results: Synchronized oscillations in GFP expression were repeatedly observed in chemically linked sub-populations of cells. Computational simulations predicted that the introduction of independently inducible repressors substantially broaden the range of conditions under which oscillations could occur, in addition to allowing the frequency of the oscillation to be tuned.
Conclusions: The genetic circuits described here may prove to be valuable research tools for the study of synchronized transcriptional feedback loops under a variety of conditions and experimental set-ups. We further demonstrate the benefit of using abstract visualizations to discover subtle non-linear trends in complex dynamic models with large parameter spaces.
Figures


References
-
- Hasty J, McMillen D, Isaacs F, Collins JJ. Computational studies of gene regulatory networks: in numero molecular biology. Nat Rev Genet. 2001;2(4):268–279. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

