Do plants contain g protein-coupled receptors?
- PMID: 24246381
- PMCID: PMC3875809
- DOI: 10.1104/pp.113.228874
Do plants contain g protein-coupled receptors?
Abstract
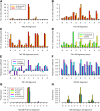
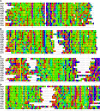
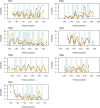
Whether G protein-coupled receptors (GPCRs) exist in plants is a fundamental biological question. Interest in deorphanizing new GPCRs arises because of their importance in signaling. Within plants, this is controversial, as genome analysis has identified 56 putative GPCRs, including G protein-coupled receptor1 (GCR1), which is reportedly a remote homolog to class A, B, and E GPCRs. Of these, GCR2 is not a GPCR; more recently, it has been proposed that none are, not even GCR1. We have addressed this disparity between genome analysis and biological evidence through a structural bioinformatics study, involving fold recognition methods, from which only GCR1 emerges as a strong candidate. To further probe GCR1, we have developed a novel helix-alignment method, which has been benchmarked against the class A-class B-class F GPCR alignments. In addition, we have presented a mutually consistent set of alignments of GCR1 homologs to class A, class B, and class F GPCRs and shown that GCR1 is closer to class A and/or class B GPCRs than class A, class B, or class F GPCRs are to each other. To further probe GCR1, we have aligned transmembrane helix 3 of GCR1 to each of the six GPCR classes. Variability comparisons provide additional evidence that GCR1 homologs have the GPCR fold. From the alignments and a GCR1 comparative model, we have identified motifs that are common to GCR1, class A, B, and E GPCRs. We discuss the possibilities that emerge from this controversial evidence that GCR1 has a GPCR fold.
Figures

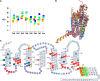
References
-
- Attwood TK, Findlay JB. (1994) Fingerprinting G-protein-coupled receptors. Protein Eng 7: 195–203 - PubMed
-
- Baldwin JM, Schertler GF, Unger VM. (1997) An alpha-carbon template for the transmembrane helices in the rhodopsin family of G-protein-coupled receptors. J Mol Biol 272: 144–164 - PubMed
-
- Ballesteros JA, Weinstein H. (1995) Integrated methods for the construction of three-dimensional models and computational probing of structure-function relationships in G-protein coupled receptors. Methods Neurosci 25: 366–428
-
- Bennett-Lovsey RM, Herbert AD, Sternberg MJ, Kelley LA. (2008) Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins 70: 611–625 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

