Detectable clonal mosaicism in the human genome
- PMID: 24246702
- PMCID: PMC3855860
- DOI: 10.1053/j.seminhematol.2013.09.001
Detectable clonal mosaicism in the human genome
Abstract
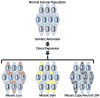
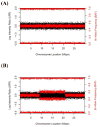
Human genetic mosaicism is the presence of two or more cellular populations with distinct genotypes in an individual who developed from a single fertilized ovum. While initially observed across a spectrum of rare genetic disorders, detailed assessment of data from genome-wide association studies now reveal that detectable clonal mosaicism involving large structural alterations (>2 Mb) can also be seen in populations of apparently healthy individuals. The first generation of descriptive studies has generated new interest in understanding the molecular basis of the affected genomic regions, percent of the cellular subpopulation involved, and developmental timing of the underlying mutational event, which could reveal new insights into the initiation, clonal expansion, and phenotypic manifestations of mosaic events. Early evidence indicates detectable clonal mosaicism increases in frequency with age and could preferentially occur in males. The observed pattern of recurrent events affecting specific chromosomal regions indicates some regions are more susceptible to these events, which could reflect inter-individual differences in genomic stability. Moreover, it is also plausible that the presence of large structural events could be associated with cancer risk. The characterization of detectable genetic mosaicism reveals that there could be important dynamic changes in the human genome associated with the aging process, which could be associated with risk for common disorders, such as cancer, cardiovascular disease, diabetes, and neurological disorders.
© 2013 Published by Elsevier Inc.
Figures



References
-
- Zlotnikoff M. A Human Mosaic. J Heredity. 1945;36:163–8.
-
- Strachan T, Read AP, Strachan T. Human molecular genetics. 4. New York: Garland Science; 2011. p. xxv.p. 781.
-
- Larsson NG. Somatic mitochondrial DNA mutations in mammalian aging. Annu Rev Biochem. 2010;79:683–706. - PubMed
-
- Bianchi NO, Bianchi MS, Richard SM. Mitochondrial genome instability in human cancers. Mutat Res. 2001;488:9–23. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

