Mammalian protein arginine methyltransferase 7 (PRMT7) specifically targets RXR sites in lysine- and arginine-rich regions
- PMID: 24247247
- PMCID: PMC3873558
- DOI: 10.1074/jbc.M113.525345
Mammalian protein arginine methyltransferase 7 (PRMT7) specifically targets RXR sites in lysine- and arginine-rich regions
Abstract
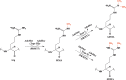
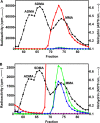
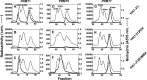
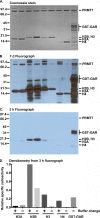
The mammalian protein arginine methyltransferase 7 (PRMT7) has been implicated in roles of transcriptional regulation, DNA damage repair, RNA splicing, cell differentiation, and metastasis. However, the type of reaction that it catalyzes and its substrate specificity remain controversial. In this study, we purified a recombinant mouse PRMT7 expressed in insect cells that demonstrates a robust methyltransferase activity. Using a variety of substrates, we demonstrate that the enzyme only catalyzes the formation of ω-monomethylarginine residues, and we confirm its activity as the prototype type III protein arginine methyltransferase. This enzyme is active on all recombinant human core histones, but histone H2B is a highly preferred substrate. Analysis of the specific methylation sites within intact histone H2B and within H2B and H4 peptides revealed novel post-translational modification sites and a unique specificity of PRMT7 for methylating arginine residues in lysine- and arginine-rich regions. We demonstrate that a prominent substrate recognition motif consists of a pair of arginine residues separated by one residue (RXR motif). These findings will significantly accelerate substrate profile analysis, biological function study, and inhibitor discovery for PRMT7.
Keywords: Histone Methylation; Mass Spectrometry (MS); Methyltransferases; Post-translational Modification; Protein Methylation; Protein-arginine Methyltransferases (PRMT); S-Adenosylmethionine (AdoMet).
Figures





Similar articles
-
Human protein arginine methyltransferase 7 (PRMT7) is a type III enzyme forming ω-NG-monomethylated arginine residues.J Biol Chem. 2012 Mar 9;287(11):7859-70. doi: 10.1074/jbc.M111.336271. Epub 2012 Jan 12. J Biol Chem. 2012. PMID: 22241471 Free PMC article.
-
Substrate specificity of human protein arginine methyltransferase 7 (PRMT7): the importance of acidic residues in the double E loop.J Biol Chem. 2014 Nov 21;289(47):32604-16. doi: 10.1074/jbc.M114.609271. Epub 2014 Oct 7. J Biol Chem. 2014. PMID: 25294873 Free PMC article.
-
The macromolecular complexes of histones affect protein arginine methyltransferase activities.J Biol Chem. 2021 Oct;297(4):101123. doi: 10.1016/j.jbc.2021.101123. Epub 2021 Sep 6. J Biol Chem. 2021. PMID: 34492270 Free PMC article.
-
PRMT7 as a unique member of the protein arginine methyltransferase family: A review.Arch Biochem Biophys. 2019 Apr 15;665:36-45. doi: 10.1016/j.abb.2019.02.014. Epub 2019 Feb 22. Arch Biochem Biophys. 2019. PMID: 30802433 Free PMC article. Review.
-
Non-Histone Arginine Methylation by Protein Arginine Methyltransferases.Curr Protein Pept Sci. 2020;21(7):699-712. doi: 10.2174/1389203721666200507091952. Curr Protein Pept Sci. 2020. PMID: 32379587 Free PMC article. Review.
Cited by
-
Arginine Methylation of the PGC-1α C-Terminus Is Temperature-Dependent.Biochemistry. 2023 Jan 3;62(1):22-34. doi: 10.1021/acs.biochem.2c00363. Epub 2022 Dec 19. Biochemistry. 2023. PMID: 36535003 Free PMC article.
-
Independent transcriptomic and proteomic regulation by type I and II protein arginine methyltransferases.iScience. 2021 Aug 11;24(9):102971. doi: 10.1016/j.isci.2021.102971. eCollection 2021 Sep 24. iScience. 2021. PMID: 34505004 Free PMC article.
-
Caenorhabditis elegans PRMT-7 and PRMT-9 Are Evolutionarily Conserved Protein Arginine Methyltransferases with Distinct Substrate Specificities.Biochemistry. 2017 May 23;56(20):2612-2626. doi: 10.1021/acs.biochem.7b00283. Epub 2017 May 9. Biochemistry. 2017. PMID: 28441492 Free PMC article.
-
Discovery of four recessive developmental disorders using probabilistic genotype and phenotype matching among 4,125 families.Nat Genet. 2015 Nov;47(11):1363-9. doi: 10.1038/ng.3410. Epub 2015 Oct 5. Nat Genet. 2015. PMID: 26437029 Free PMC article.
-
A comprehensive review of histone modifications during mammalian oogenesis and early embryo development.Histochem Cell Biol. 2025 Jun 28;163(1):70. doi: 10.1007/s00418-025-02398-x. Histochem Cell Biol. 2025. PMID: 40580240 Free PMC article. Review.
References
-
- Walsh C. T. (2006) Post-translational Modification of Proteins: Expanding Nature's Inventory, pp. 1–490, Roberts and Co. Publishers, Englewood, CO
-
- Yang Y., Bedford M. T. (2013) Protein-arginine methyltransferases and cancer. Nat. Rev. Cancer 13, 37–50 - PubMed
-
- Miranda T. B., Miranda M., Frankel A., Clarke S. (2004) PRMT7 is a member of the protein arginine methyltransferase family with a distinct substrate specificity. J. Biol. Chem. 279, 22902–22907 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

