SAXS structural study of PrP(Sc) reveals ~11 nm diameter of basic double intertwined fibers
- PMID: 24247356
- PMCID: PMC4201618
- DOI: 10.4161/pri.27190
SAXS structural study of PrP(Sc) reveals ~11 nm diameter of basic double intertwined fibers
Abstract
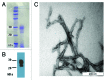
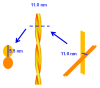
A sample of purified Syrian hamster PrP27-30 prion fibers was analyzed by synchrotron small-angle X-ray scattering (SAXS). The SAXS pattern obtained was fitted to a model based on infinitely long cylinders with a log-normal intensity distribution, a hard-sphere structure factor and a general Porod term for larger aggregates. The diameter calculated for the cylinders determined from the fit was 11.0 ± 0.2 nm. This measurement offers an estimation of the diameter of PrP(Sc) fibers in suspension, i.e., free of errors derived from estimations based on 2D projections in transmission electron microscopy images, subjected to further possible distortions from the negative stain. This diameter, which corresponds to a maximum diameter of approximately 5.5 nm for each of the two intertwined protofilaments making up the fibers, rules out the possibility that PrP(Sc) conforms to a stack of in-register, single-rung flat PrP(Sc) monomers; rather, PrP(Sc) subunits must necessarily coil, most likely several times, into themselves.
Keywords: PrPSc structure; SAXS; TEM; amyloid; prion; synchrotron radiation.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
