Studies of translational misreading in vivo show that the ribosome very efficiently discriminates against most potential errors
- PMID: 24249223
- PMCID: PMC3866648
- DOI: 10.1261/rna.039792.113
Studies of translational misreading in vivo show that the ribosome very efficiently discriminates against most potential errors
Abstract
Protein synthesis must rapidly and repeatedly discriminate between a single correct and many incorrect aminoacyl-tRNAs. We have attempted to measure the frequencies of all possible missense errors by tRNA , tRNA and tRNA . The most frequent errors involve three types of mismatched nucleotide pairs, U•U, U•C, or U•G, all of which can form a noncanonical base pair with geometry similar to that of the canonical U•A or C•G Watson-Crick pairs. Our system is sensitive enough to measure errors at other potential mismatches that occur at frequencies as low as 1 in 500,000 codons. The ribosome appears to discriminate this efficiently against any pair with non-Watson-Crick geometry. This extreme accuracy may be necessary to allow discrimination against the errors involving near Watson-Crick pairing.
Keywords: Escherichia coli; mistranslation; non-Watson–Crick base pairs; protein synthesis; β-galactosidase.
Figures
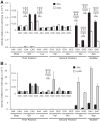
 on mutants of codon 537 vary by over two orders of magnitude. The activity of mutants with the indicated codons in place of Glu codon 537 are shown relative to the activity of wild-type β-galactosidase (error bars, SEM). The insets use greatly expanded y-axis to represent the much lower activities of the 10 indicated mutants. (A) Comparison of mutant activities in the wild-type (XAc) and hyperaccurate (rpsL) genetic backgrounds. (B) Comparison of activities in wild-type and error-prone (rpsD) backgrounds.
on mutants of codon 537 vary by over two orders of magnitude. The activity of mutants with the indicated codons in place of Glu codon 537 are shown relative to the activity of wild-type β-galactosidase (error bars, SEM). The insets use greatly expanded y-axis to represent the much lower activities of the 10 indicated mutants. (A) Comparison of mutant activities in the wild-type (XAc) and hyperaccurate (rpsL) genetic backgrounds. (B) Comparison of activities in wild-type and error-prone (rpsD) backgrounds.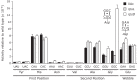
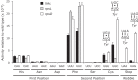

 , the frequency of misreading could not be more than 4 × 10–4 per codon. The arrows are labeled to indicate the position of the mismatch and color coded for the nature of the mismatched base pair, as shown.
, the frequency of misreading could not be more than 4 × 10–4 per codon. The arrows are labeled to indicate the position of the mismatch and color coded for the nature of the mismatched base pair, as shown.References
-
- Andersson DI, Bohman K, Isaksson LA, Kurland CG 1982. Translation rates and misreading characteristics of rpsD mutants in Escherichia coli. Mol Gen Genet 187: 467–472 - PubMed
-
- Dahlgren A, Ryden-Aulin M 2000. A novel mutation in ribosomal protein S4 that affects the function of a mutated RF1. Biochimie 82: 683–691 - PubMed
-
- Demeshkina N, Jenner L, Westhof E, Yusupov M, Yusupova G 2012. A new understanding of the decoding principle on the ribosome. Nature 484: 256–259 - PubMed
-
- Dong H, Nilsson L, Kurland CG 1996. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J Mol Biol 260: 649–663 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
