Biophysical characterization of G-quadruplex forming FMR1 mRNA and of its interactions with different fragile X mental retardation protein isoforms
- PMID: 24249225
- PMCID: PMC3866639
- DOI: 10.1261/rna.041442.113
Biophysical characterization of G-quadruplex forming FMR1 mRNA and of its interactions with different fragile X mental retardation protein isoforms
Abstract
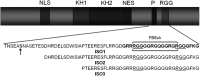
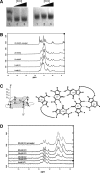
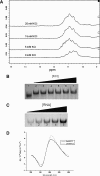
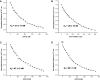
Fragile X syndrome, the most common form of inherited mental impairment in humans, is caused by the absence of the fragile X mental retardation protein (FMRP) due to a CGG trinucleotide repeat expansion in the 5'-untranslated region (UTR) and subsequent translational silencing of the fragile x mental retardation-1 (FMR1) gene. FMRP, which is proposed to be involved in the translational regulation of specific neuronal messenger RNA (mRNA) targets, contains an arginine-glycine-glycine (RGG) box RNA binding domain that has been shown to bind with high affinity to G-quadruplex forming mRNA structures. FMRP undergoes alternative splicing, and the binding of FMRP to a proposed G-quadruplex structure in the coding region of its mRNA (named FBS) has been proposed to affect the mRNA splicing events at exon 15. In this study, we used biophysical methods to directly demonstrate the folding of FMR1 FBS into a secondary structure that contains two specific G-quadruplexes and analyze its interactions with several FMRP isoforms. Our results show that minor splice isoforms, ISO2 and ISO3, created by the usage of the second and third acceptor sites at exon 15, bind with higher affinity to FBS than FMRP ISO1, which is created by the usage of the first acceptor site. FMRP ISO2 and ISO3 cannot undergo phosphorylation, an FMRP post-translational modification shown to modulate the protein translation regulation. Thus, their expression has to be tightly regulated, and this might be accomplished by a feedback mechanism involving the FMRP interactions with the G-quadruplex structures formed within FMR1 mRNA.
Keywords: FMRP; G-quadruplex RNA; fluorescence spectroscopy; protein–RNA interactions.
Figures


References
-
- Ashley CT, Sutcliffe JS, Kunst CB, Leiner HA, Eichler EE, Nelson DL, Warren ST 1993. Human and murine FMR-1: Alternative splicing and translational initiation downstream of the CGG-repeat. Nat Genet 4: 244–251 - PubMed
-
- Bole M, Menon L, Mihailescu MR 2008. Fragile X mental retardation protein recognition of G quadruplex structure per se is sufficient for high affinity binding to RNA. Mol Biosyst 4: 1212–1219 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
