tRNA 3' processing in yeast involves tRNase Z, Rex1, and Rrp6
- PMID: 24249226
- PMCID: PMC3866640
- DOI: 10.1261/rna.041467.113
tRNA 3' processing in yeast involves tRNase Z, Rex1, and Rrp6
Abstract
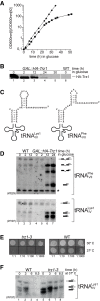
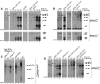
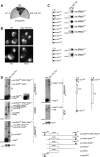
Mature tRNA 3' ends in the yeast Saccharomyces cerevisiae are generated by two pathways: endonucleolytic and exonucleolytic. Although two exonucleases, Rex1 and Rrp6, have been shown to be responsible for the exonucleolytic trimming, the identity of the endonuclease has been inferred from other systems but not confirmed in vivo. Here, we show that the yeast tRNA 3' endonuclease tRNase Z, Trz1, is catalyzing endonucleolytic tRNA 3' processing. The majority of analyzed tRNAs utilize both pathways, with a preference for the endonucleolytic one. However, 3'-end processing of precursors with long 3' trailers depends to a greater extent on Trz1. In addition to its function in the nucleus, Trz1 processes the 3' ends of mitochondrial tRNAs, contributing to the general RNA metabolism in this organelle.
Keywords: 3′-end processing; Saccharomyces cerevisiae; exonucleases; tRNA; tRNase Z.
Figures




References
-
- Arends S, Schön A 1997. Partial purification and characterization of nuclear ribonuclease P from wheat. Eur J Biochem 244: 635–645 - PubMed
-
- Benschop JJ, Brabers N, van Leenen D, Bakker LV, van Deutekom HW, van Berkum NL, Apweiler E, Lijnzaad P, Holstege FC, Kemmeren P 2010. A consensus of core protein complex compositions for Saccharomyces cerevisiae. Mol Cell 38: 916–928 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
