Enzymatic production of mono-ubiquitinated proteins for structural studies: The example of the Josephin domain of ataxin-3
- PMID: 24251111
- PMCID: PMC3829987
- DOI: 10.1016/j.fob.2013.10.005
Enzymatic production of mono-ubiquitinated proteins for structural studies: The example of the Josephin domain of ataxin-3
Abstract
Protein ubiquitination occurs through formation of an isopeptide bond between the C-terminal glycine of ubiquitin (Ub) and the ɛ-amino group of a substrate lysine residue. This post-translational modification, which occurs through the attachment of single and/or multiple copies of mono-ubiquitin and poly-ubiquitin chains, is involved in crucial cellular events such as protein degradation, cell-cycle regulation and DNA repair. The abnormal functioning of ubiquitin pathways is also implicated in the pathogenesis of several human diseases ranging from cancer to neurodegeneration. However, despite the undoubted biological importance, understanding the molecular basis of how ubiquitination regulates different pathways has up to now been strongly limited by the difficulty of producing the amounts of highly homogeneous samples that are needed for a structural characterization by X-ray crystallography and/or NMR. Here, we report on the production of milligrams of highly pure Josephin mono-ubiquitinated on lysine 117 through large scale in vitro enzymatic ubiquitination. Josephin is the catalytic domain of ataxin-3, a protein responsible for spinocerebellar ataxia type 3. Ataxin-3 is the first deubiquitinating enzyme (DUB) reported to be activated by mono-ubiquitination. We demonstrate that the samples produced with the described method are correctly folded and suitable for structural studies. The protocol allows facile selective labelling of the components. Our results provide an important proof-of-concept that may pave the way to new approaches to the in vitro study of ubiquitinated proteins.
Keywords: ATP, adenosine triphosphate; DTT, dithiothreitol; DUB, deubiquitinating enzyme; Deubiquitinating enzyme; GST, glutathione-S-transferase; HSQC, heteronuclear single quantum coherence; IAA, iodoacetamide; Isopeptide bond; JosK117-only, Josephin mutant in which all lysines but K117 are mutated; Josephin; MS/MS tandem, mass spectrometry; Machado–Joseph disease; NMR, nuclear magnetic resonance; PDB, Protein Data Bank; Post-translational modification; SDS–PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis; Spinocerebellar ataxia type 3; Tris–HCl, 2-amino-2-(hydroxymethyl)-1,3-propanediol hydrochloride; Ubiquitin.
Figures

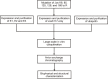
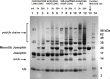


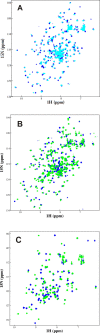
References
-
- Goldknopf I.L., Taylor C.W., Baum R.M., Yeoman L.C., Olson M.O., Prestayko A.W., Busch H. Isolation and characterization of protein A24, a “histone-like” non-histone chromosomal protein. J. Biol. Chem. 1975;250:7182–7187. - PubMed
-
- Sadowski M., Suryadinata R., Tan A.R., Roesley S.N., Sarcevic B. Protein monoubiquitination and polyubiquitination generate structural diversity to control distinct biological processes. IUBMB Life. 2012;64:136–142. - PubMed
-
- Komander D., Rape M. The ubiquitin code. Annu. Rev. Biochem. 2012;81:203–229. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

