Molecular epidemiology of Rabbit Haemorrhagic Disease Virus in Australia: when one became many
- PMID: 24251353
- PMCID: PMC4595043
- DOI: 10.1111/mec.12596
Molecular epidemiology of Rabbit Haemorrhagic Disease Virus in Australia: when one became many
Abstract
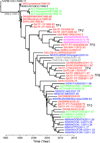
Rabbit Haemorrhagic Disease Virus (RHDV) was introduced into Australia in 1995 as a biological control agent against the wild European rabbit (Oryctolagus cuniculus). We evaluated its evolution over a 16-year period (1995-2011) by examining 50 isolates collected throughout Australia, as well as the original inoculum strains. Phylogenetic analysis of capsid protein VP60 sequences of the Australian isolates, compared with those sampled globally, revealed that they form a monophyletic group with the inoculum strains (CAPM V-351 and RHDV351INOC). Strikingly, despite more than 3000 rereleases of RHDV351INOC since 1995, only a single viral lineage has sustained its transmission in the long-term, indicative of a major competitive advantage. In addition, we find evidence for widespread viral gene flow, in which multiple lineages entered individual geographic locations, resulting in a marked turnover of viral lineages with time, as well as a continual increase in viral genetic diversity. The rate of RHDV evolution recorded in Australia -4.0 (3.3-4.7) × 10(-3) nucleotide substitutions per site per year - was higher than previously observed in RHDV, and evidence for adaptive evolution was obtained at two VP60 residues. Finally, more intensive study of a single rabbit population (Turretfield) in South Australia provided no evidence for viral persistence between outbreaks, with genetic diversity instead generated by continual strain importation.
Keywords: European rabbit; Rabbit Haemorrhagic Disease Virus; biocontrol; epidemiology; evolution; phylogeny.
© 2013 John Wiley & Sons Ltd.
Figures



References
-
- Abrantes J, Esteves PJ, van der Loo W. Evidence for recombination in the major capsid gene VP60 of the rabbit haemorrhagic disease virus (RHDV) Archives of Virology. 2008;153:329–335. - PubMed
-
- Asgari S, Hardy JR, Sinclair RG, Cooke BD. Field evidence for mechanical transmission of rabbit haemorrhagic disease virus (RHDV) by flies (Diptera: Calliphoridae) among wild rabbits in Australia. Virus Research. 1998;54:123–132. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

