Oxidative stress and inflammation modulate Rev-erbα signaling in the neonatal lung and affect circadian rhythmicity
- PMID: 24252172
- PMCID: PMC4048579
- DOI: 10.1089/ars.2013.5539
Oxidative stress and inflammation modulate Rev-erbα signaling in the neonatal lung and affect circadian rhythmicity
Abstract
Aims: The response to oxidative stress and inflammation varies with diurnal rhythms. Nevertheless, it is not known whether circadian genes are regulated by these stimuli. We evaluated whether Rev-erbα, a key circadian gene, was regulated by oxidative stress and/or inflammation in vitro and in a mouse model.
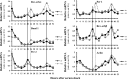
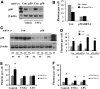
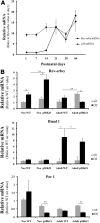
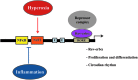
Results: A unique sequence consisting of overlapping AP-1 and nuclear factor kappa B (NFκB) consensus sequences was identified on the mouse Rev-erbα promoter. This sequence mediates Rev-erbα promoter activity and transcription in response to oxidative stress and inflammation. This region serves as an NrF2 platform both to receive oxidative stress signals and to activate Rev-erbα, as well as an NFκB-binding site to repress Rev-erbα with inflammatory stimuli. The amplitude of the rhythmicity of Rev-erbα was altered by pre-exposure to hyperoxia or disruption of NFκB in a cell culture model of circadian simulation. Oxidative stress overcame the inhibitory effect of NFκB binding on Rev-erbα transcription. This was confirmed in neonatal mice exposed to hyperoxia, where hyperoxia-induced lung Rev-erbα transcription was further increased with NFκB disruption. Interestingly, this effect was not observed in similarly exposed adult mice.
Innovation: These data provide novel mechanistic insights into how key circadian genes are regulated by oxidative stress and inflammation in the neonatal lung.
Conclusion: Rev-erbα transcription and circadian oscillation are susceptible to oxidative stress and inflammation in the neonate. Due to Rev-erbα's role in cellular metabolism, this could contribute to lung cellular function and injury from inflammation and oxidative stress.
Figures






Similar articles
-
The nuclear receptor Rev-erbα participates in circadian regulation of Ugt2b enzymes in mice.Biochem Pharmacol. 2019 Mar;161:89-97. doi: 10.1016/j.bcp.2019.01.010. Epub 2019 Jan 11. Biochem Pharmacol. 2019. PMID: 30639455
-
Circadian clock protein Rev-erbα regulates neuroinflammation.Proc Natl Acad Sci U S A. 2019 Mar 12;116(11):5102-5107. doi: 10.1073/pnas.1812405116. Epub 2019 Feb 21. Proc Natl Acad Sci U S A. 2019. PMID: 30792350 Free PMC article.
-
REV-ERBα alters circadian rhythms by modulating mTOR signaling.Mol Cell Endocrinol. 2021 Feb 5;521:111108. doi: 10.1016/j.mce.2020.111108. Epub 2020 Dec 5. Mol Cell Endocrinol. 2021. PMID: 33285244
-
Dissecting the Rev-erbα Cistrome and the Mechanisms Controlling Circadian Transcription in Liver.Cold Spring Harb Symp Quant Biol. 2015;80:233-8. doi: 10.1101/sqb.2015.80.027508. Epub 2015 Sep 14. Cold Spring Harb Symp Quant Biol. 2015. PMID: 26370410 Review.
-
The nuclear receptor REV-ERBα integrates circadian clock and energy metabolism.Yi Chuan. 2023 Feb 20;45(2):99-114. doi: 10.16288/j.yczz.22-310. Yi Chuan. 2023. PMID: 36927658 Review.
Cited by
-
The circadian gene Rev-erbα improves cellular bioenergetics and provides preconditioning for protection against oxidative stress.Free Radic Biol Med. 2016 Apr;93:177-89. doi: 10.1016/j.freeradbiomed.2016.02.004. Epub 2016 Feb 5. Free Radic Biol Med. 2016. PMID: 26855417 Free PMC article.
-
The wrinkling of time: Aging, inflammation, oxidative stress, and the circadian clock in neurodegeneration.Neurobiol Dis. 2020 Jun;139:104832. doi: 10.1016/j.nbd.2020.104832. Epub 2020 Mar 13. Neurobiol Dis. 2020. PMID: 32179175 Free PMC article. Review.
-
CeO2NPs relieve radiofrequency radiation, improve testosterone synthesis, and clock gene expression in Leydig cells by enhancing antioxidation.Int J Nanomedicine. 2019 Jun 24;14:4601-4611. doi: 10.2147/IJN.S206561. eCollection 2019. Int J Nanomedicine. 2019. PMID: 31296989 Free PMC article.
-
NF-κB modifies the mammalian circadian clock through interaction with the core clock protein BMAL1.PLoS Genet. 2021 Nov 22;17(11):e1009933. doi: 10.1371/journal.pgen.1009933. eCollection 2021 Nov. PLoS Genet. 2021. PMID: 34807912 Free PMC article.
-
Targeting Circadian Protein Rev-erbα to Alleviate Inflammation, Oxidative Stress, and Enhance Functional Recovery Following Brain Trauma.Antioxidants (Basel). 2024 Jul 25;13(8):901. doi: 10.3390/antiox13080901. Antioxidants (Basel). 2024. PMID: 39199147 Free PMC article.
References
-
- Angelov D, Lenouvel F, Hans F, Muller CW, Bouvet P, Bednar J, Moudrianakis EN, Cadet J, and Dimitrov S. The histone octamer is invisible when NF-kappaB binds to the nucleosome. J Biol Chem 279: 42374–42382, 2004 - PubMed
-
- Balsalobre A, Damiola F, and Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93: 929–937, 1998 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
