TTG2-regulated development is related to expression of putative AUXIN RESPONSE FACTOR genes in tobacco
- PMID: 24252253
- PMCID: PMC4046668
- DOI: 10.1186/1471-2164-14-806
TTG2-regulated development is related to expression of putative AUXIN RESPONSE FACTOR genes in tobacco
Abstract
Background: The phytohormone auxin mediates a stunning array of plant development through the functions of AUXIN RESPONSE FACTORs (ARFs), which belong to transcription factors and are present as a protein family comprising 10-43 members so far identified in different plant species. Plant development is also subject to regulation by TRANSPARENT TESTA GLABRA (TTG) proteins, such as NtTTG2 that we recently characterized in tobacco Nicotiana tabacum. To find the functional linkage between TTG and auxin in the regulation of plant development, we performed de novo assembly of the tobacco transcriptome to identify candidates of NtTTG2-regulated ARF genes.
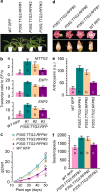
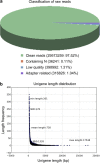
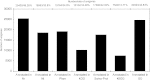
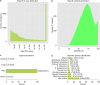
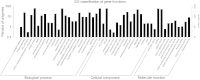
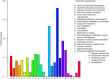
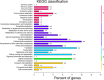
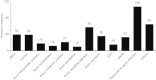
Results: The role of NtTTG2 in tobacco growth and development was studied by analyzing the biological effects of gene silencing and overexpression. The NtTTG2 gene silencing causes repressive effects on vegetative growth, floral anthocyanin synthesis, flower colorization, and seed production. By contrast, the plant growth and development processes are promoted by NtTTG2 overexpression. The growth/developmental function of NtTTG2 associates with differential expression of putative ARF genes identified by de novo assembly of the tobacco transcriptome. The transcriptome contains a total of 54,906 unigenes, including 30,124 unigenes (54.86%) with annotated functions and at least 8,024 unigenes (14.61%) assigned to plant growth and development. The transcriptome also contains 455 unigenes (0.83%) related to auxin responses, including 40 putative ARF genes. Based on quantitative analyses, the expression of the putative genes is either promoted or inhibited by NtTTG2.
Conclusions: The biological effects of the NtTTG2 gene silencing and overexpression suggest that NtTTG2 is an essential regulator of growth and development in tobacco. The effects of the altered NtTTG2 expression on expression levels of putative ARF genes identified in the transcriptome suggest that NtTTG2 functions in relation to ARF transcription factors.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

