A high-density SNP genetic linkage map for the silver-lipped pearl oyster, Pinctada maxima: a valuable resource for gene localisation and marker-assisted selection
- PMID: 24252414
- PMCID: PMC4046678
- DOI: 10.1186/1471-2164-14-810
A high-density SNP genetic linkage map for the silver-lipped pearl oyster, Pinctada maxima: a valuable resource for gene localisation and marker-assisted selection
Abstract
Background: The silver-lipped pearl oyster, Pinctada maxima, is an important tropical aquaculture species extensively farmed for the highly sought "South Sea" pearls. Traditional breeding programs have been initiated for this species in order to select for improved pearl quality, but many economic traits under selection are complex, polygenic and confounded with environmental factors, limiting the accuracy of selection. The incorporation of a marker-assisted selection (MAS) breeding approach would greatly benefit pearl breeding programs by allowing the direct selection of genes responsible for pearl quality. However, before MAS can be incorporated, substantial genomic resources such as genetic linkage maps need to be generated. The construction of a high-density genetic linkage map for P. maxima is not only essential for unravelling the genomic architecture of complex pearl quality traits, but also provides indispensable information on the genome structure of pearl oysters.
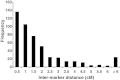
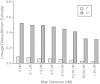
Results: A total of 1,189 informative genome-wide single nucleotide polymorphisms (SNPs) were incorporated into linkage map construction. The final linkage map consisted of 887 SNPs in 14 linkage groups, spans a total genetic distance of 831.7 centimorgans (cM), and covers an estimated 96% of the P. maxima genome. Assessment of sex-specific recombination across all linkage groups revealed limited overall heterochiasmy between the sexes (i.e. 1.15:1 F/M map length ratio). However, there were pronounced localised differences throughout the linkage groups, whereby male recombination was suppressed near the centromeres compared to female recombination, but inflated towards telomeric regions. Mean values of LD for adjacent SNP pairs suggest that a higher density of markers will be required for powerful genome-wide association studies. Finally, numerous nacre biomineralization genes were localised providing novel positional information for these genes.
Conclusions: This high-density SNP genetic map is the first comprehensive linkage map for any pearl oyster species. It provides an essential genomic tool facilitating studies investigating the genomic architecture of complex trait variation and identifying quantitative trait loci for economically important traits useful in genetic selection programs within the P. maxima pearling industry. Furthermore, this map provides a foundation for further research aiming to improve our understanding of the dynamic process of biomineralization, and pearl oyster evolution and synteny.
Figures






Similar articles
-
Genome-wide SNP validation and mantle tissue transcriptome analysis in the silver-lipped pearl oyster, Pinctada maxima.Mar Biotechnol (NY). 2013 Dec;15(6):647-58. doi: 10.1007/s10126-013-9514-3. Epub 2013 May 30. Mar Biotechnol (NY). 2013. PMID: 23715808
-
In silico whole-genome EST analysis reveals 2322 novel microsatellites for the silver-lipped pearl oyster, Pinctada maxima.Mar Genomics. 2011 Dec;4(4):287-90. doi: 10.1016/j.margen.2011.06.007. Epub 2011 Aug 15. Mar Genomics. 2011. PMID: 22118641
-
An Expressed Sequence Tag (EST)-enriched genetic map of turbot (Scophthalmus maximus): a useful framework for comparative genomics across model and farmed teleosts.BMC Genet. 2012 Jul 2;13:54. doi: 10.1186/1471-2156-13-54. BMC Genet. 2012. PMID: 22747677 Free PMC article.
-
Aquaculture genomics, genetics and breeding in the United States: current status, challenges, and priorities for future research.BMC Genomics. 2017 Feb 20;18(1):191. doi: 10.1186/s12864-017-3557-1. BMC Genomics. 2017. PMID: 28219347 Free PMC article. Review.
-
Current knowledge in lentil genomics and its application for crop improvement.Front Plant Sci. 2015 Feb 23;6:78. doi: 10.3389/fpls.2015.00078. eCollection 2015. Front Plant Sci. 2015. PMID: 25755659 Free PMC article. Review.
Cited by
-
Construction of the First High-Density Genetic Linkage Map and Analysis of Quantitative Trait Loci for Growth-Related Traits in Sinonovacula constricta.Mar Biotechnol (NY). 2017 Oct;19(5):488-496. doi: 10.1007/s10126-017-9768-2. Epub 2017 Jul 19. Mar Biotechnol (NY). 2017. PMID: 28725940
-
Genomic Tools and Selective Breeding in Molluscs.Front Genet. 2018 Jul 18;9:253. doi: 10.3389/fgene.2018.00253. eCollection 2018. Front Genet. 2018. PMID: 30073016 Free PMC article. Review.
-
Sex Differences in the Recombination Landscape.Am Nat. 2020 Feb;195(2):361-379. doi: 10.1086/704943. Epub 2019 Dec 9. Am Nat. 2020. PMID: 32017625 Free PMC article.
-
Sex Differences in Recombination in Sticklebacks.G3 (Bethesda). 2018 May 31;8(6):1971-1983. doi: 10.1534/g3.118.200166. G3 (Bethesda). 2018. PMID: 29632132 Free PMC article.
-
Genetic mapping and QTL analysis of growth-related traits in Pinctada fucata using restriction-site associated DNA sequencing.PLoS One. 2014 Nov 4;9(11):e111707. doi: 10.1371/journal.pone.0111707. eCollection 2014. PLoS One. 2014. PMID: 25369421 Free PMC article.
References
-
- Tisdell C, Poirine B. Economics of pearl farming. In: Southgate PC, Lucas JS, editors. The pearl oyster. Amsterdam, The Netherlands: Elsevier B V; 2008. pp. 473–495.
-
- Wada KT, Jerry DR. Population genetics and stock improvement. In: Southgate PC, Lucas JS, editors. The pearl oyster. Amsterdam, The Netherlands: Elsevier B V; 2008. pp. 437–471.
-
- Kvingedal R, Evans BS, Lind CE, Taylor JJU, Dupont-Nivet M, Jerry DR. Population and family growth response to different rearing location, heritability estimates and genotype × environment interaction in the silver-lip pearl oyster (Pinctada maxima) Aquaculture. 2010;304(1–4):1–6. doi: 10.1016/j.aquaculture.2010.02.035. - DOI
-
- Jerry DR, Kvingedal R, Lind CE, Evans BS, Taylor JJU, Safari AE. Donor-oyster derived heritability estimates and the effect of genotype x environment interaction on the production of pearl quality traits in the silver-lip pearl oyster, Pinctada maxima. Aquaculture. 2012;338–341:66–71. doi: 10.1016/j.aquaculture.2012.02.001. - DOI
-
- Saavedra C, Bachere E. Bivalve genomics. Aquaculture. 2006;256(1–4):1–14. doi: 10.1016/j.aquaculture.2006.02.023. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

