The DNA methyltransferase Dnmt1 directly interacts with the SET and RING finger-associated (SRA) domain of the multifunctional protein Uhrf1 to facilitate accession of the catalytic center to hemi-methylated DNA
- PMID: 24253042
- PMCID: PMC3879560
- DOI: 10.1074/jbc.M113.523209
The DNA methyltransferase Dnmt1 directly interacts with the SET and RING finger-associated (SRA) domain of the multifunctional protein Uhrf1 to facilitate accession of the catalytic center to hemi-methylated DNA
Abstract
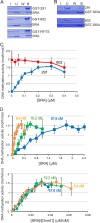
Dnmt1 is responsible for the maintenance DNA methylation during replication to propagate methylation patterns to the next generation. The replication foci targeting sequence (RFTS), which plugs the catalytic pocket, is necessary for recruitment of Dnmt1 to the replication site. In the present study we found that the DNA methylation activity of Dnmt1 was DNA length-dependent and scarcely methylated 12-bp short hemi-methylated DNA. Contrarily, the RFTS-deleted Dnmt1 and Dnmt1 mutants that destroyed the hydrogen bonds between the RFTS and catalytic domain showed significant DNA methylation activity even toward 12-bp hemi-methylated DNA. The DNA methylation activity of the RFTS-deleted Dnmt1 toward 12-bp hemi-methylated DNA was strongly inhibited on the addition of RFTS, but to a lesser extent by Dnmt1 harboring the mutations that impair the hydrogen bond formation. The SRA domain of Uhrf1, which is a prerequisite factor for maintenance methylation and selectively binds to hemi-methylated DNA, stimulated the DNA methylation activity of Dnmt1. The SRA to Dnmt1 concentration ratio was the determinant for the maximum stimulation. In addition, a mutant SRA, which had lost the DNA binding activity but was able to bind to Dnmt1, stimulated the DNA methylation activity of Dnmt1. The results indicate that the DNA methylation activity of Dnmt1 was stimulated on the direct interaction of the SRA and Dnmt1. The SRA facilitated acceptance of the 12-bp fluorocytosine-containing DNA by the catalytic center. We propose that the SRA removes the RFTS plug from the catalytic pocket to facilitate DNA acceptance by the catalytic center.
Keywords: DNA Methylation; DNA Methyltransferase; Epigenetics; Gene Regulation; Gene Silencing.
Figures




References
-
- Li E. (2002) Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 3, 662–673 - PubMed
-
- Reik W., Lewis A. (2005) Co-evolution of X chromosome inactivation and imprinting in mammals. Nat. Rev. Genet. 6, 403–410 - PubMed
-
- Goll M. G., Bestor T. H. (2005) Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 74, 481–514 - PubMed
-
- Suetake I., Hayata D., Tajima S. (2006) The amino terminus of mouse DNA methyltransferase 1 forms an independent domain and binds to DNA with the sequence involving PCNA binding motif. J. Biochem. 140, 763–776 - PubMed
-
- Rountree M. R., Bachman K. E., Baylin S. B. (2000) DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat. Genet. 25, 269–277 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

