New insights into aluminum tolerance in rice: the ASR5 protein binds the STAR1 promoter and other aluminum-responsive genes
- PMID: 24253199
- PMCID: PMC3973494
- DOI: 10.1093/mp/sst160
New insights into aluminum tolerance in rice: the ASR5 protein binds the STAR1 promoter and other aluminum-responsive genes
Abstract
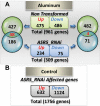
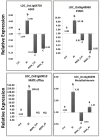
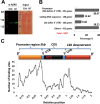
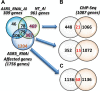
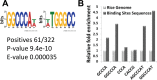
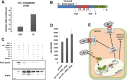
Aluminum (Al) toxicity in plants is one of the primary constraints in crop production. Al³⁺, the most toxic form of Al, is released into soil under acidic conditions and causes extensive damage to plants, especially in the roots. In rice, Al tolerance requires the ASR5 gene, but the molecular function of ASR5 has remained unknown. Here, we perform genome-wide analyses to identify ASR5-dependent Al-responsive genes in rice. Based on ASR5_RNAi silencing in plants, a global transcriptome analysis identified a total of 961 genes that were responsive to Al treatment in wild-type rice roots. Of these genes, 909 did not respond to Al in the ASR5_RNAi plants, indicating a central role for ASR5 in Al-responsive gene expression. Under normal conditions, without Al treatment, the ASR5_RNAi plants expressed 1.756 genes differentially compared to the wild-type plants, and 446 of these genes responded to Al treatment in the wild-type plants. Chromatin immunoprecipitation followed by deep sequencing identified 104 putative target genes that were directly regulated by ASR5 binding to their promoters, including the STAR1 gene, which encodes an ABC transporter required for Al tolerance. Motif analysis of the binding peak sequences revealed the binding motif for ASR5, which was confirmed via in vitro DNA-binding assays using the STAR1 promoter. These results demonstrate that ASR5 acts as a key transcription factor that is essential for Al-responsive gene expression and Al tolerance in rice.
Keywords: ASR.; Aluminum; ChIP-Seq; RNA-Seq; rice.
Figures

References
-
- Arenhart R.A, Lima J.C de, Pedron M., Carvalho F.E.L., Silveira J.A., Rosa S.B., Caverzan A., Andrade C.M.B., Schünemann M., Margis R., et al. (2013). Involvement of ASR genes in aluminium tolerance mechanisms in rice. Plant, Cell & Environment. 36, 52–67 - PubMed
-
- Baier A.C., Somers D.J., Gusiafson J.P. (1995). Aluminium tolerance in wheat: correlating hydroponic evaluations with field and soil performances. Plant Breeding. 114, 291–296
-
- Barcelo J., Poschenrieder C. (2002). Fast root growth responses, root exudates, and internal detoxification as clues to the mechanisms of aluminium toxicity and resistance: a review. Environmental and Experimental Botany. 48, 75–92
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

