Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases
- PMID: 24253446
- PMCID: PMC3875854
- DOI: 10.1101/gr.162339.113
Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases
Abstract
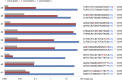
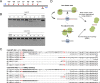
RNA-guided endonucleases (RGENs), derived from the prokaryotic adaptive immune system known as CRISPR/Cas, enable targeted genome engineering in cells and organisms. RGENs are ribonucleoproteins that consist of guide RNA and Cas9, a protein component originated from Streptococcus pyogenes. These enzymes cleave chromosomal DNA, whose sequence is complementary, to guide RNA in a targeted manner, producing site-specific DNA double-strand breaks (DSBs), the repair of which gives rise to targeted genome modifications. Despite broad interest in RGEN-mediated genome editing, these nucleases are limited by off-target mutations and unwanted chromosomal translocations associated with off-target DNA cleavages. Here, we show that off-target effects of RGENs can be reduced below the detection limits of deep sequencing by choosing unique target sequences in the genome and modifying both guide RNA and Cas9. We found that both the composition and structure of guide RNA can affect RGEN activities in cells to reduce off-target effects. RGENs efficiently discriminated on-target sites from off-target sites that differ by two bases. Furthermore, exome sequencing analysis showed that no off-target mutations were induced by two RGENs in four clonal populations of mutant cells. In addition, paired Cas9 nickases, composed of D10A Cas9 and guide RNA, which generate two single-strand breaks (SSBs) or nicks on different DNA strands, were highly specific in human cells, avoiding off-target mutations without sacrificing genome-editing efficiency. Interestingly, paired nickases induced chromosomal deletions in a targeted manner without causing unwanted translocations. Our results highlight the importance of choosing unique target sequences and optimizing guide RNA and Cas9 to avoid or reduce RGEN-induced off-target mutations.
Figures





References
-
- Bae KH, Kwon YD, Shin HC, Hwang MS, Ryu EH, Park KS, Yang HY, Lee DK, Lee Y, Park J, et al. 2003. Human zinc fingers as building blocks in the construction of artificial transcription factors. Nat Biotechnol 21: 275–280 - PubMed
-
- Bibikova M, Beumer K, Trautman JK, Carroll D 2003. Enhancing gene targeting with designed zinc finger nucleases. Science 300: 764. - PubMed
-
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U 2009. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326: 1509–1512 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
