Analogue encoding of physicochemical properties of proteins in their cognate messenger RNAs
- PMID: 24253588
- PMCID: PMC3868254
- DOI: 10.1038/ncomms3784
Analogue encoding of physicochemical properties of proteins in their cognate messenger RNAs
Abstract
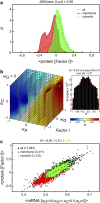
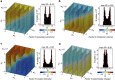
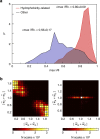
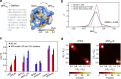
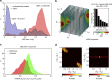
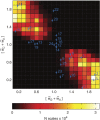
Being related by the genetic code, mRNAs and their cognate proteins exhibit mutually interdependent compositions, which implies the possibility of a direct connection between their general physicochemical properties. Here we probe the general potential of the cell to encode information about proteins in the average characteristics of their cognate mRNAs and decode it in a ribosome-independent manner. We show that average protein hydrophobicity, calculated from either sequences or 3D structures, can be encoded in an analogue fashion by many different average mRNA sequence properties with the only constraint being that pyrimidine and purine bases be clearly distinguishable on average. Moreover, average characteristics of mRNA sequences enable discrimination between cytosolic and membrane proteins even in the absence of topogenic signal-based mechanisms. Our results suggest that protein and mRNA localization may be partly determined by basic physicochemical rationales and interdependencies between the two biomolecules.
Figures
References
-
- McLane L. M. & Corbett A. H. Nuclear localization signals and human disease. IUBMB Life 61, 697–706 (2009). - PubMed
-
- Verkman A. S. Solute and macromolecule diffusion in cellular aqueous compartments. Trends Biochem. Sci. 27, 27–33 (2002). - PubMed
-
- Brangwynne C. P., Koenderink G. H., MacKintosh F. C. & Weitz D. A. Intracellular transport by active diffusion. Trends Cell Biol. 19, 423–427 (2009). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

