cor, a novel carbon monoxide resistance gene, is essential for Mycobacterium tuberculosis pathogenesis
- PMID: 24255121
- PMCID: PMC3870250
- DOI: 10.1128/mBio.00721-13
cor, a novel carbon monoxide resistance gene, is essential for Mycobacterium tuberculosis pathogenesis
Abstract
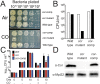
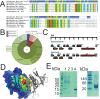
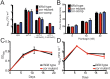
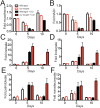
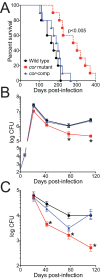
Tuberculosis, caused by Mycobacterium tuberculosis, remains a devastating human infectious disease, causing two million deaths annually. We previously demonstrated that M. tuberculosis induces an enzyme, heme oxygenase (HO1), that produces carbon monoxide (CO) gas and that M. tuberculosis adapts its transcriptome during CO exposure. We now demonstrate that M. tuberculosis carries a novel resistance gene to combat CO toxicity. We screened an M. tuberculosis transposon library for CO-susceptible mutants and found that disruption of Rv1829 (carbon monoxide resistance, Cor) leads to marked CO sensitivity. Heterologous expression of Cor in Escherichia coli rescued it from CO toxicity. Importantly, the virulence of the cor mutant is attenuated in a mouse model of tuberculosis. Thus, Cor is necessary and sufficient to protect bacteria from host-derived CO. Taken together, this represents the first report of a role for HO1-derived CO in controlling infection of an intracellular pathogen and the first identification of a CO resistance gene in a pathogenic organism.
Importance: Macrophages produce a variety of antimicrobial molecules, including nitric oxide (NO), hydrogen peroxide (H2O2), and acid (H+), that serve to kill engulfed bacteria. In addition to these molecules, human and mouse macrophages also produce carbon monoxide (CO) gas by the heme oxygenase (HO1) enzyme. We observed that, in contrast to other bacteria, mycobacteria are resistant to CO, suggesting that this might be an evolutionary adaptation of mycobacteria for survival within macrophages. We screened a panel of ~2,500 M. tuberculosis mutants to determine which genes are required for survival of M. tuberculosis in the presence of CO. Within this panel, we identified one such gene, cor, that specifically confers CO resistance. Importantly, we found that the ability of M. tuberculosis cells carrying a mutated copy of this gene to cause tuberculosis in a mouse disease model is significantly attenuated. This indicates that CO resistance is essential for mycobacterial survival in vivo.
Figures

References
-
- Huynh KK, Joshi SA, Brown EJ. 2011. A delicate dance: host response to mycobacteria. Curr. Opin. Immunol. 23:464–472 - PubMed
-
- Liu PT, Modlin RL. 2008. Human macrophage host defense against Mycobacterium tuberculosis. Curr. Opin. Immunol. 20:371–376 - PubMed
-
- Rohde K, Yates RM, Purdy GE, Russell DG. 2007. Mycobacterium tuberculosis and the environment within the phagosome. Immunol. Rev. 219:37–54 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 5T32AI007520/AI/NIAID NIH HHS/United States
- T32 AI060537/AI/NIAID NIH HHS/United States
- AI099439/AI/NIAID NIH HHS/United States
- R56 AI081727/AI/NIAID NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- T32 AI007520/AI/NIAID NIH HHS/United States
- K08 AI076632/AI/NIAID NIH HHS/United States
- P01 AI063302/AI/NIAID NIH HHS/United States
- R01 AI099439/AI/NIAID NIH HHS/United States
- AI081727/AI/NIAID NIH HHS/United States
- DK081668/DK/NIDDK NIH HHS/United States
- AI076632/AI/NIAID NIH HHS/United States
- K08 DK081668/DK/NIDDK NIH HHS/United States
- R01 AI081727/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

