Characterization of RNA in exosomes secreted by human breast cancer cell lines using next-generation sequencing
- PMID: 24255815
- PMCID: PMC3828613
- DOI: 10.7717/peerj.201
Characterization of RNA in exosomes secreted by human breast cancer cell lines using next-generation sequencing
Abstract
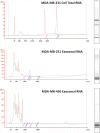
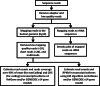
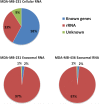
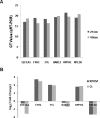
Exosomes are nanosized (30-100 nm) membrane vesicles secreted by most cell types. Exosomes have been found to contain various RNA species including miRNA, mRNA and long non-protein coding RNAs. A number of cancer cells produce elevated levels of exosomes. Because exosomes have been isolated from most body fluids they may provide a source for non-invasive cancer diagnostics. Transcriptome profiling that uses deep-sequencing technologies (RNA-Seq) offers enormous amount of data that can be used for biomarkers discovery, however, in case of exosomes this approach was applied only for the analysis of small RNAs. In this study, we utilized RNA-Seq technology to analyze RNAs present in microvesicles secreted by human breast cancer cell lines. Exosomes were isolated from the media conditioned by two human breast cancer cell lines, MDA-MB-231 and MDA-MB-436. Exosomal RNA was profiled using the Ion Torrent semiconductor chip-based technology. Exosomes were found to contain various classes of RNA with the major class represented by fragmented ribosomal RNA (rRNA), in particular 28S and 18S rRNA subunits. Analysis of exosomal RNA content revealed that it reflects RNA content of the donor cells. Although exosomes produced by the two cancer cell lines shared most of the RNA species, there was a number of non-coding transcripts unique to MDA-MB-231 and MDA-MB-436 cells. This suggests that RNA analysis might distinguish exosomes produced by low metastatic breast cancer cell line (MDA-MB-436) from that produced by highly metastatic breast cancer cell line (MDA-MB-231). The analysis of gene ontologies (GOs) associated with the most abundant transcripts present in exosomes revealed significant enrichment in genes encoding proteins involved in translation and rRNA and ncRNA processing. These GO terms indicate most expressed genes for both, cellular and exosomal RNA. For the first time, using RNA-seq, we examined the transcriptomes of exosomes secreted by human breast cancer cells. We found that most abundant exosomal RNA species are the fragments of 28S and 18S rRNA subunits. This limits the number of reads from other RNAs. To increase the number of detectable transcripts and improve the accuracy of their expression level the protocols allowing depletion of fragmented rRNA should be utilized in the future RNA-seq analyses on exosomes. Present data revealed that exosomal transcripts are representative of their cells of origin and thus could form basis for detection of tumor specific markers.
Keywords: Biomarkers; Breast cancer; Exosomes; Microvesicles; Next generation sequencing.
Figures






References
-
- Bobrie A, Krumeich S, Reyal F, Recchi C, Moita LF, Seabra MC, Ostrowski M, Thery C. Rab27a supports exosome-dependent and -independent mechanisms that modify the tumor microenvironment and can promote tumor progression. Cancer Res. 2012;72:4920–4930. doi: 10.1158/0008-5472.CAN-12-0925. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

