Moenomycin resistance mutations in Staphylococcus aureus reduce peptidoglycan chain length and cause aberrant cell division
- PMID: 24255971
- PMCID: PMC3944067
- DOI: 10.1021/cb4006744
Moenomycin resistance mutations in Staphylococcus aureus reduce peptidoglycan chain length and cause aberrant cell division
Abstract
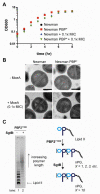
Staphylococcus aureus is a Gram-positive pathogen with an unusual mode of cell division in that it divides in orthogonal rather than parallel planes. Through selection using moenomycin, an antibiotic proposed to target peptidoglycan glycosyltransferases (PGTs), we have generated resistant mutants containing a single point mutation in the active site of the PGT domain of an essential peptidoglycan (PG) biosynthetic enzyme, PBP2. Using cell free polymerization assays, we show that this mutation alters PGT activity so that much shorter PG chains are made. The same mutation in another S. aureus PGT, SgtB, has a similar effect on glycan chain length. Moenomycin-resistant S. aureus strains containing mutated PGTs that make only short glycan polymers display major cell division defects, implicating PG chain length in determining bacterial cell morphology and division site placement.
Figures


References
-
- Vollmer W, Blanot D, de Pedro MA. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 2008;32:149–167. - PubMed
-
- Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 2008;32:234–258. - PubMed
-
- Lovering AL, Gretes M, Strynadka NCJ. Structural details of the glycosyltransferase step of peptidoglycan assembly. Curr. Opin. Struct. Biol. 2008;18:534–543. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

