The RNA- and DNA-targeting CRISPR-Cas immune systems of Pyrococcus furiosus
- PMID: 24256230
- PMCID: PMC3996508
- DOI: 10.1042/BST20130056
The RNA- and DNA-targeting CRISPR-Cas immune systems of Pyrococcus furiosus
Abstract
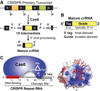
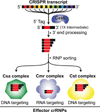
Using the hyperthermophile Pyrococcus furiosus, we have delineated several key steps in CRISPR (clustered regularly interspaced short palindromic repeats)-Cas (CRISPR-associated) invader defence pathways. P. furiosus has seven transcriptionally active CRISPR loci that together encode a total of 200 crRNAs (CRISPR RNAs). The 27 Cas proteins in this organism represent three distinct pathways and are primarily encoded in two large gene clusters. The Cas6 protein dices CRISPR locus transcripts to generate individual invader-targeting crRNAs. The mature crRNAs include a signature sequence element (the 5' tag) derived from the CRISPR locus repeat sequence that is important for function. crRNAs are tailored into distinct species and integrated into three distinct crRNA-Cas protein complexes that are all candidate effector complexes. The complex formed by the Cmr [Cas module RAMP (repeat-associated mysterious proteins)] (subtype III-B) proteins cleaves complementary target RNAs and can be programmed to cleave novel target RNAs in a prokaryotic RNAi-like manner. Evidence suggests that the other two CRISPR-Cas systems in P. furiosus, Csa (Cas subtype Apern) (subtype I-A) and Cst (Cas subtype Tneap) (subtype I-B), target invaders at the DNA level. Studies of the CRISPR-Cas systems from P. furiosus are yielding fundamental knowledge of mechanisms of crRNA biogenesis and silencing for three of the diverse CRISPR-Cas pathways, and reveal that organisms such as P. furiosus possess an arsenal of multiple RNA-guided mechanisms to resist diverse invaders. Our knowledge of the fascinating CRISPR-Cas pathways is leading in turn to our ability to co-opt these systems for exciting new biomedical and biotechnological applications.
Figures

References
-
- Sorek R, Lawrence CM, Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu. Rev. Biochem. 2013;82:237–266. - PubMed
-
- Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. - PubMed
-
- Westra ER, Swarts DC, Staals RH, Jore MM, Brouns SJ, van der Oost J. The CRISPRs, they are a-changin’: how prokaryotes generate adaptive immunity. Annu. Rev. Genet. 2012;46:311–339. - PubMed
-
- Bhaya D, Davison M, Barrangou R. CRISPR–Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 2011;45:273–297. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

