Generation of “LYmph Node Derived Antibody Libraries” (LYNDAL) for selecting fully human antibody fragments with therapeutic potential
- PMID: 24256717
- PMCID: PMC3929437
- DOI: 10.4161/mabs.27236
Generation of “LYmph Node Derived Antibody Libraries” (LYNDAL) for selecting fully human antibody fragments with therapeutic potential
Abstract
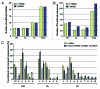
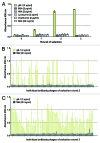
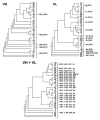
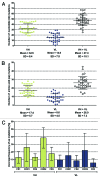
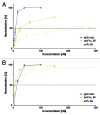
The development of efficient strategies for generating fully human monoclonal antibodies with unique functional properties that are exploitable for tailored therapeutic interventions remains a major challenge in the antibody technology field. Here, we present a methodology for recovering such antibodies from antigen-encountered human B cell repertoires. As the source for variable antibody genes, we cloned immunoglobulin G (IgG)-derived B cell repertoires from lymph nodes of 20 individuals undergoing surgery for head and neck cancer. Sequence analysis of unselected “LYmph Node Derived Antibody Libraries” (LYNDAL) revealed a naturally occurring distribution pattern of rearranged antibody sequences, representing all known variable gene families and most functional germline sequences. To demonstrate the feasibility for selecting antibodies with therapeutic potential from these repertoires, seven LYNDAL from donors with high serum titers against herpes simplex virus (HSV) were panned on recombinant glycoprotein B of HSV-1. Screening for specific binders delivered 34 single-chain variable fragments (scFvs) with unique sequences. Sequence analysis revealed extensive somatic hypermutation of enriched clones as a result of affinity maturation. Binding of scFvs to common glycoprotein B variants from HSV-1 and HSV-2 strains was highly specific, and the majority of analyzed antibody fragments bound to the target antigen with nanomolar affinity. From eight scFvs with HSV-neutralizing capacity in vitro,the most potent antibody neutralized 50% HSV-2 at 4.5 nM as a dimeric (scFv)2. We anticipate our approach to be useful for recovering fully human antibodies with therapeutic potential.
Figures




References
-
- Köhler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. 1975. J Immunol. 2005;174:2453–5. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
